PCR special primer pair for identifying pig-cattle-sheep derived ingredients in livestock and poultry meat based on mitochondrion COI gene, PCR identification method and reagent kit
An identification method, a technology of source components, applied in the field of food testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
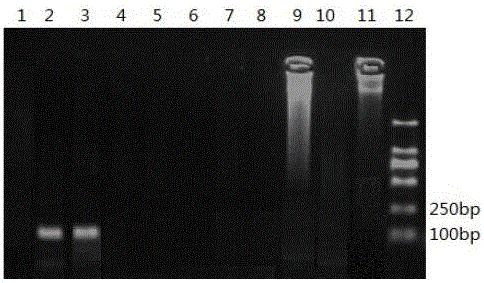
[0051] Extraction of total muscle DNA of various species: fully mix fresh pork, beef, mutton, rabbit, pigeon, quail, chicken, duck and goose respectively, and use the above centrifugal column tissue genomic DNA extraction kit DNA was extracted, and the extracted total DNA samples were dissolved in 100ul of TE eluate, detected by 1% agarose gel electrophoresis, and the absorbance values at 260 nm and 280 nm were measured by a nucleic acid protein analyzer, and the DNA concentration and purity were calculated, and stored at -20 °C spare.
[0052] Design of specific primers: According to the gene sequences of pig, cattle, sheep, rabbit, pigeon, quail, chicken, duck and goose mitochondrial DNA COⅠ published in GenBank, and analyzed by BLAST, PrimerPremier5 was used for each species-specific base site. .0 software screened out porcine, bovine and sheep specific primer pairs. Each primer is described below:
[0053] Primer pair I for amplifying the porcine COⅠ gene to generate p...
Embodiment 2
[0063] Taking the total DNA samples of each species in Example 1 as a template:
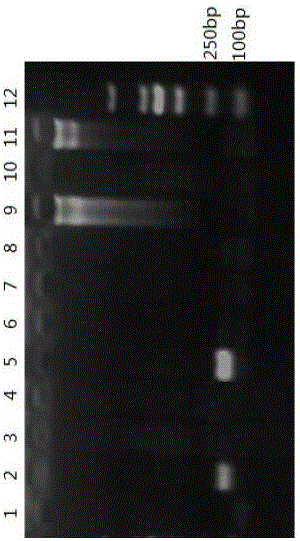
[0064] (1) Using the three pairs of primers in Example 1, a PCR detection system was established using pork, beef, and mutton DNA as templates, other animal muscle DNA as a control, and double-distilled sterilized water without nucleic acid as a negative control.
[0065] (2) Preparation of 20 μL reaction system: 2×PCR Mix 10ul, 10 μmol / L forward primer pair I or primer pair II or primer pair III 0.3 μL, 10 μmol / L reverse primer pair I or primer pair II or primer pair III 0.3 μL , template 1 μL, double-distilled sterilized water 8.4 μL.
[0066] (3) The formulation of the PCR reaction program: firstly, pre-denaturation at 95°C for 5 min; then denaturation at 95°C for 30s, annealing at 60°C for 30s, and extension at 72°C for 60s for 30 cycles; and finally extension at 72°C for 10 min.
[0067] (4) Agarose gel electrophoresis: Take the PCR amplification product obtained in (4) and detect it by 1.5...
Embodiment 3
[0073] Using the mixed total DNA samples of 9 animals as a template:
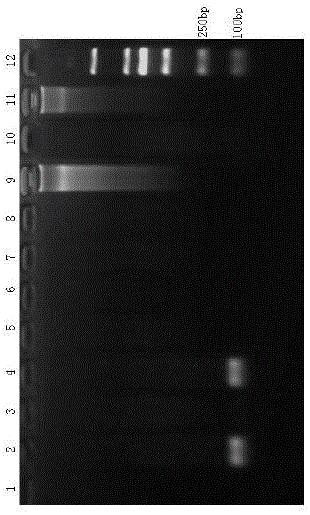
[0074] (1) Treatment of mixed total DNA samples: Mix equal volumes of the nine animal total DNA samples in Example 1, wherein each animal muscle DNA occupies 0.1 volume, and mix with 0.1 volume of double-distilled sterilized water as Mix DNA templates.
[0075] (2) The three pairs of primers in Example 1 were respectively used, with mixed DNA as template, 9 animal muscle DNAs as control, and double-distilled sterilized water without nucleic acid as negative control, to establish a PCR detection system.
[0076] (3) Preparation of 20 μL reaction system: 2×PCR Mix 10ul, 10 μmol / L forward primer pair I or primer pair II or primer pair III 0.3 μL, 10 μmol / L reverse primer pair I or primer pair II or primer pair III 0.3 μL , mix 1 μL of DNA template and 8.4 μL of double-distilled sterilized water.
[0077] (4) Formulation of PCR reaction program: The reaction program is the same as the reaction program in Exam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com