Method for synthesizing 3-O-glucose-based oleanolic acid and cellobiose oleanolic acid by using saccharomyces cerevisiae
A kind of Saccharomyces cerevisiae and gene technology, applied in the field of bioengineering, can solve the problems such as the difficult realization of chemical synthesis of molecular structure, and achieve the effects of increasing production, optimizing fermentation process, and reducing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Synthetic Genes CYP716A12, MtCPR and UGT73C10
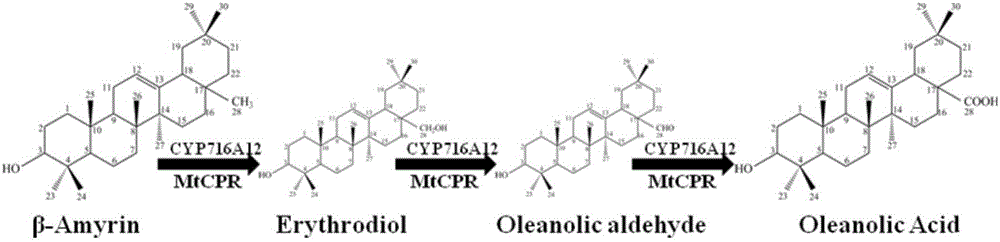
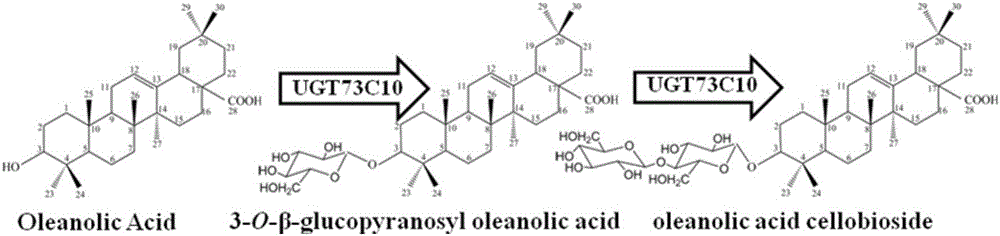
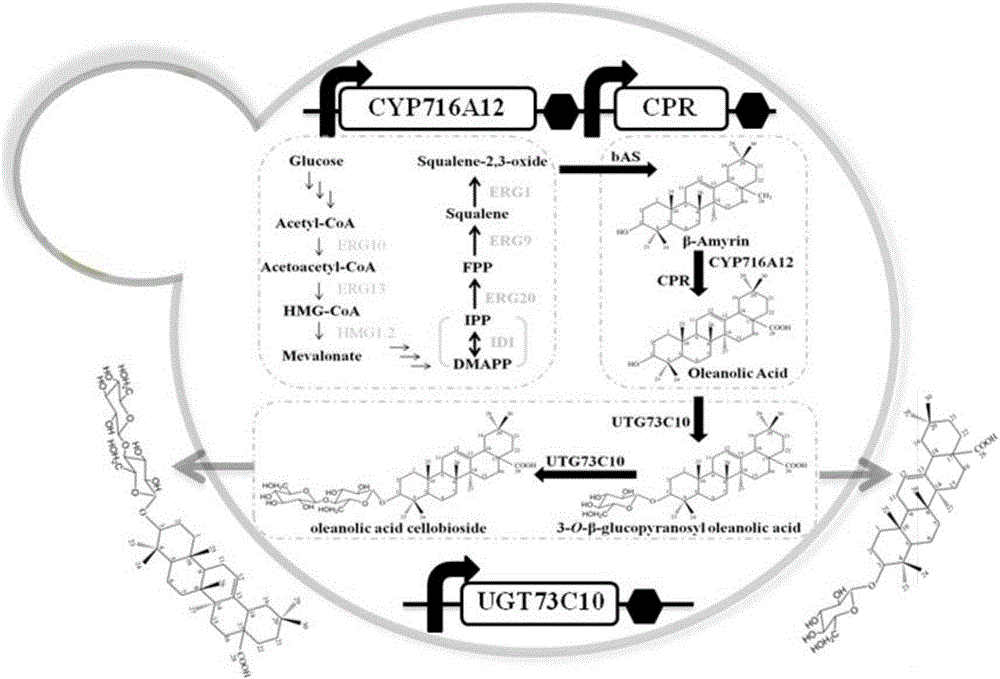
[0022] The P450 cytochrome monooxygenase encoded by the gene CYP716A12 and the cytochrome reductase encoded by the gene MtCPR can work together to oxidize β-amyresinol to oleanolic acid (see figure 1 ); the UDP-glucotransferase encoded by the gene UGT73C10 can glycosylate the hydroxyl group at the C-3 position of oleanolic acid to generate 3-O-glucosyl oleanolic acid and cellobiose oleanolic acid (see figure 2 ). The P450 cytochrome monooxygenase gene CYP716A12 from Medicago truncatula (Medicago truncatula), the cytochrome reductase gene MtCPR from Medicago truncatula (Medicago truncatula) and the P450 cytochrome reductase gene MtCPR from European mountain mustard (Barbarea vulgaris) were retrieved from the NCBI database. For the sequence of UDP-glucose transferase gene UGT73C10, the codon optimization software JCAT was used to optimize the codons of the three genes, so that the amino acid coding sequence confo...
Embodiment 2
[0023] Example 2 Construction of expression vectors for genes CYP716A12, MtCPR and UGT73C10
[0024] Using the codon-optimized pUC57-CYP716A12 synthesized by the company as a template, primers CYP716A12-F-BamHI and CYP716A12-R-SalI were used to amplify the P450 cytochrome monooxygenase gene CYP716A12 of Medicago truncatula, A CYP716A12 gene fragment with a length of 1455bp was obtained. The gene fragment and the vector pESC-URA were digested simultaneously by restriction endonucleases BamHI and SalI, the gene fragment obtained above was connected to the expression vector pESC-URA, transformed into Escherichia coli TOP10 strain, and the plasmid pESC-URA-P was extracted GAL1 -CYP716A12-T CYC1 , Sequencing verification of the CYP716A12 gene on the plasmid, its sequence is shown in SEQ ID No.1, and the results show that it is correct.
[0025] Using the codon-optimized pUC57-CPR synthesized by the company as a template, the primers MtCPR-F-ClaI and MtCPR-R-NotI were used to ampl...
Embodiment 3
[0027] Example 3: Construction of engineered Saccharomyces cerevisiae producing oleanolic acid, 3-O-glucosyl oleanolic acid and cellobiose oleanolic acid
[0028] The expression vector pESC-URA-P constructed in Example 2 was transformed by lithium acetate chemical conversion method GAL1 -CYP716A12-T CYC1 -P GAL10 -MtCPR-T ADH1 and pESC-TRP-P GAL1 -UGT73C10-T CYC1 , co-transformed into S. cerevisiae strain Sgib capable of producing β-amyresinol. Spread the transformation product on a screening plate containing 2% glucose basal medium (SD-URA-TRP) lacking uracil and tryptophan, and culture at 30°C for 2-5 days. Randomly select 6-10 clones from the screening plate, and extract their plasmids respectively. Using the extracted plasmid as a template, the primer pair CYP716A12-F-BamHI and CYP716A12-R-SalI were used to amplify the P450 cytochrome monooxygenase gene CYP716A12 of Medicago truncatula; the primer pair MtCPR-F-ClaI and MtCPR-R-NotI amplifies the cytochrome reductase...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com