Rhizoctonia solani kuha SSR mark, as well as preparation method and application thereof
A corn sheath blight, marker technology, applied in the direction of biochemical equipment and methods, microorganism-based methods, DNA/RNA fragments, etc., can solve the limited, less genetic research of corn sheath blight, affecting corn yield And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Example 1: A preparation method for screening SSR markers of corn sheath blight by magnetic bead enrichment method
[0091] 1. Test materials: 60 corn sheath blight strains.
[0092] 2. Use water agar method to isolate corn sheath blight; for specific methods refer to Zhou Erxun, Yang Mei. A fast and simple technique for isolating Rhizoctonia solani from plant disease tissues. Journal of South China Agricultural University, 1998, 19(1): 125-126.
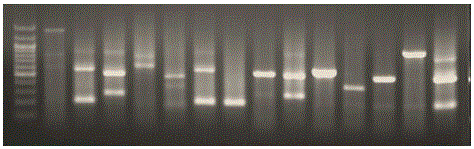
[0093] 3, get the mycelia of corn sheath blight, adopt the CTAB method to extract the total DNA of the genome; pick the mycelia of corn sheath blight and inoculate it in potato dextrose liquid medium (PDB), shake culture at constant temperature (28 ℃, 120 r / min) for 6 days, the mycelium was collected. For the specific method of extracting total genome DNA by CTAB method, refer to Huang Peitang et al., Molecular Cloning Experiment Guide (Third Edition) (Chinese translation), 2002. Take 5 μL of DNA samples for agarose gel ele...
Embodiment 2
[0142] Example 2: Application of a magnetic bead enrichment method for screening SSR markers of corn sheath blight in analyzing the genetic diversity of corn sheath blight
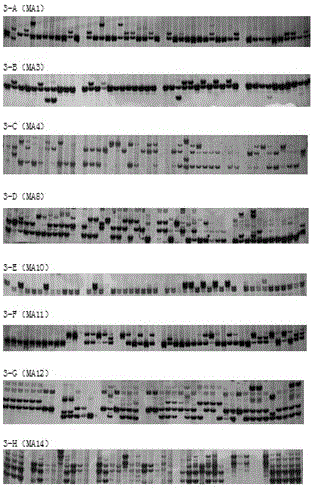
[0143] (1) Amplify 60 corn sheath blight materials with the designed SSR primers;
[0144] The test materials were all collected from the Fuyang Experimental Base of the China Rice Research Institute in Hangzhou, Zhejiang Province. Total DNA was extracted from 60 mycelia of Rhizoctonia solani and amplified by PCR. The total volume of PCR reaction is 10μL (DNA: 10ng; 10pmol / L upstream and downstream primers: 0.5μL each; 10×Taq Buffer: 1μL; 25mmol / L MgCl 2 : 1μL; 10mmol / L dNTPs: 1μL; 5U / μL Taq enzyme: 0.1μL). The PCR amplification reaction program was: pre-denaturation at 94°C for 5 min; then, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s, and 30 cycles; finally, extension at 72°C for 10 min. The PCR products were separated by molecular weight by 8% polyacrylamide g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com