Multiple PCR detection primers of escherichia coli, pseudomonas aeruginosa and staphylococcus aureus, kit and detection method
A Pseudomonas aeruginosa detection kit technology, applied in the field of microbial detection, achieves the effects of strong detection specificity, saving detection cost and time, and improving efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Design and Screening of Multiplex PCR Detection Primers
[0027] 1. Primer design
[0028] The Escherichia coli alkaline phosphatase gene (phoA gene), Pseudomonas aeruginosa outer membrane protein gene (oprL gene) and Staphylococcus aureus thermostable nuclease gene (nuc gene) were used as specific detection target sequences respectively, and primers were carried out Design and screen out 3 pairs of primers with high specificity as follows:
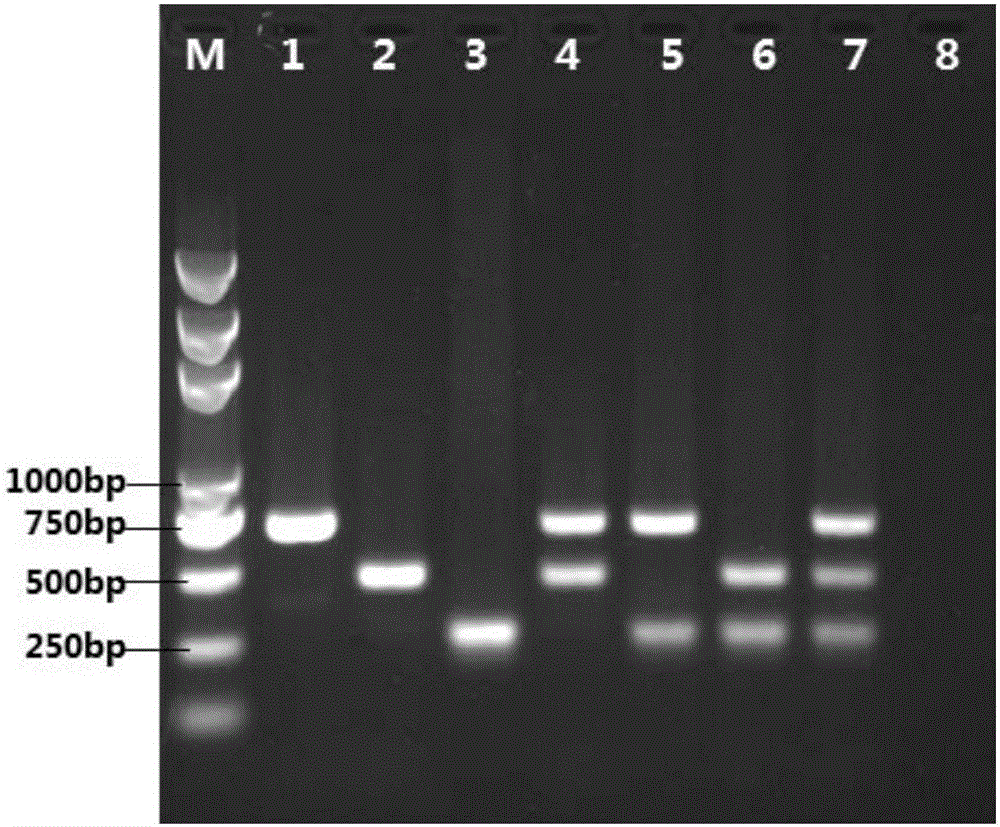
[0029] For Escherichia coli phoA gene: phoA-F: 5'-GTTTCTACCGCAGAGTTG-3' (as shown in SEQ ID NO.1), phoA-R: 5'-GACTATGACCAGCGTGTT-3' (as shown in SEQ ID NO.2), The length of the amplified fragment is 663bp.
[0030] For Pseudomonas aeruginosa oprL gene: oprL-F: 5'-GGCGTGCTGATGCTCGTA-3' (as shown in SEQ ID NO.3), oprL-R: 5'-CGCTGACCGCTGCCTTTC-3' (as shown in SEQ ID NO.4 ), the length of the amplified fragment was 444bp.
[0031] For Staphylococcus aureus nuc gene: nuc-F: 5'-ACATAAAGAACCTGCGAC-3' (as shown in SEQ ID NO....
Embodiment 2
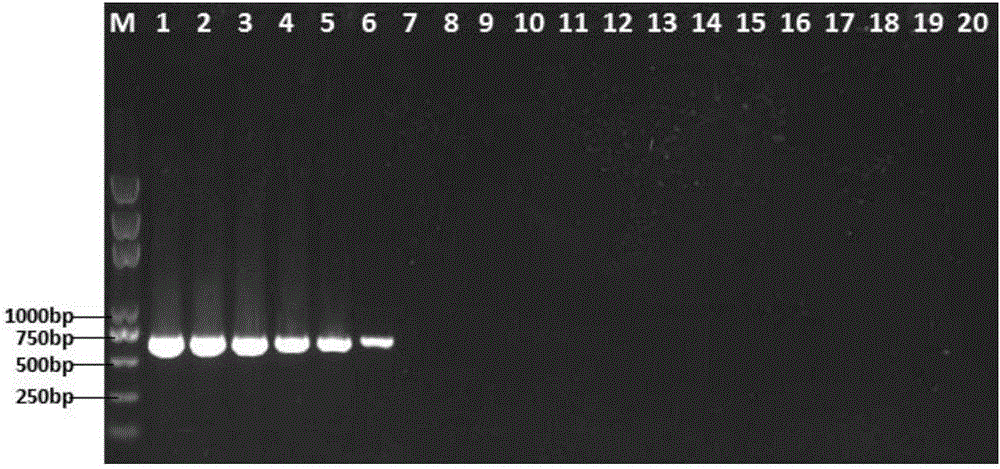
[0044] Example 2: Detection of 3 kinds of pathogenic bacteria in cosmetics
[0045] (1) Preparation of artificially polluted samples: Seven different types of cosmetic samples submitted by a company for inspection are moisturizing milk, lotion, face cream, facial cleanser, toner, liquid foundation, and essence, according to the "Hygienic Standards for Cosmetics" ( 2007 version) in the microbial detection method, no microbial contamination was found, and artificial random pathogenic bacteria contamination was carried out. Escherichia coli ATCC8039 and Pseudomonas aeruginosa ATCC9027 were added to the moisturizing milk, and Pseudomonas aeruginosa ATCC9027 and grape aureus were added to the lotion Coccus ATCC6538, Escherichia coli ATCC8039 and Staphylococcus aureus ATCC6538 were added to the cream, Escherichia coli ATCC8039, Pseudomonas aeruginosa ATCC9027 and Staphylococcus aureus ATCC6538 were added to the cleanser, Pseudomonas putida and Staphylococcus saprophyticus were added ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com