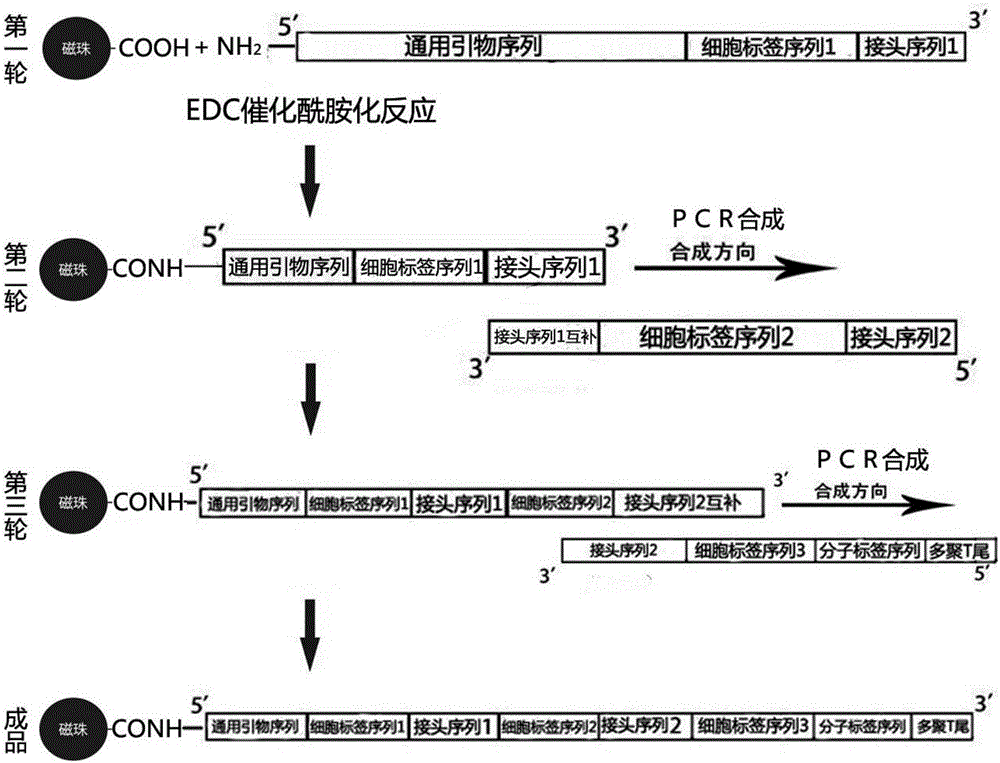
Molecular marker micro-bead and high-throughput single cell sequencing method based on same
A single-cell sequencing and molecular labeling technology, applied in the field of high-throughput single-cell sequencing and molecular-labeled microbeads, can solve the problem that single-cell sorting cannot achieve high-throughput single-cell sequencing analysis, and achieve good application value, The effect of low experimental cost and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] 1) 1ng starting DNA fragmentation
[0048]Vazyme TD513 kit was used.
[0049] a. Thaw 5×TTBL (TruePrep Tagment Buffer L) at room temperature, invert and mix well before use. Confirm that 5×TS (Terminate Solution, reaction termination solution) is at room temperature, and flick the tube wall to confirm whether there is precipitation. If there is precipitation, heat at 37°C and shake vigorously to mix well and the precipitation will dissolve.
[0050] b. Add each reaction component in sequence in the sterilized PCR tube:
[0051] 5 × TTBL 4μl
[0052] DNA 1ng
[0053] TTE Mix V1 5μl
[0054] wxya 2 O make up to 20 μl
[0055] c. Use a pipette to gently pipette 20 times to mix thoroughly.
[0056] d. Place the PCR tube in the PCR instrument and set the following reaction program:
[0057] 55°C for 10 minutes; keep warm at 10°C.
[0058] e. Immediately add 5 μl 5×TS to the reaction product, and mix well by gently blowing with a pipette. Place at room temperature ...
Embodiment 2
[0080] 1. Microplate preparation
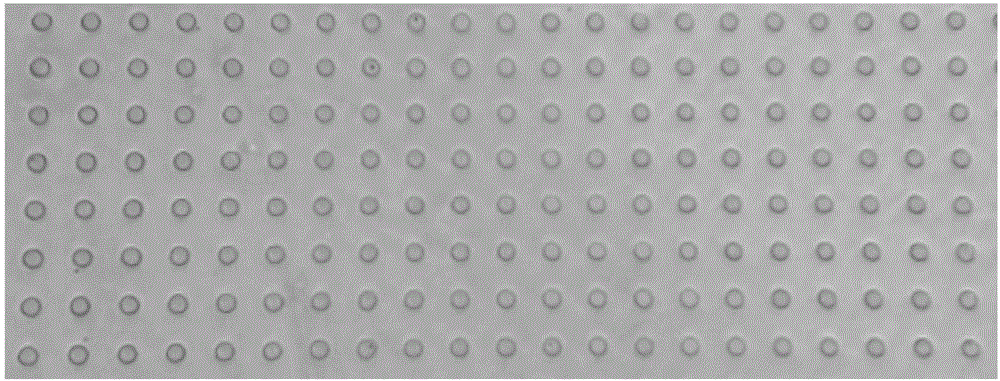
[0081] According to the experimental scale (10,000 human 293T cells and 10,000 mouse 3T3 cells each), design the size of the microwell plate (the size of the well plate is 1cm×1cm), and etch microwells on the silicon wafer as the initial mold, the depth of the microwells is 30μm and the microwell The hole size is 30 μm, and the hole spacing is 30 μm.
[0082] Next, polydimethylsiloxane (PDMS) is poured on the silicon wafer. After molding, the PDMS is taken off to become the second mold with micropillars on the plate. The microwell plate used in the final experiment is agar with a concentration of 5%. Sugar (prepared with enzyme-free water), poured on the PDMS micro-column plate to condense and form after hot-melting, and the agarose plate at this time is a micro-well plate with a certain thickness after being peeled off ( figure 1 ). When saving, add DPBS-EDTA mixture that is harmless to cells, cover and store in a 4-degree refrigerator, re...
Embodiment 3
[0116] Put 50,000 CD34 + Cells were added to a 3cm×3cm microwell plate to reach a drop-out rate of about 10%, and excess cells were washed away. Add the molecularly labeled magnetic beads into the microwell plate to achieve a hole drop rate of over 99%, and wash away the excess microwell labeled magnetic beads.
[0117] All the other steps are the same as in Example 2.
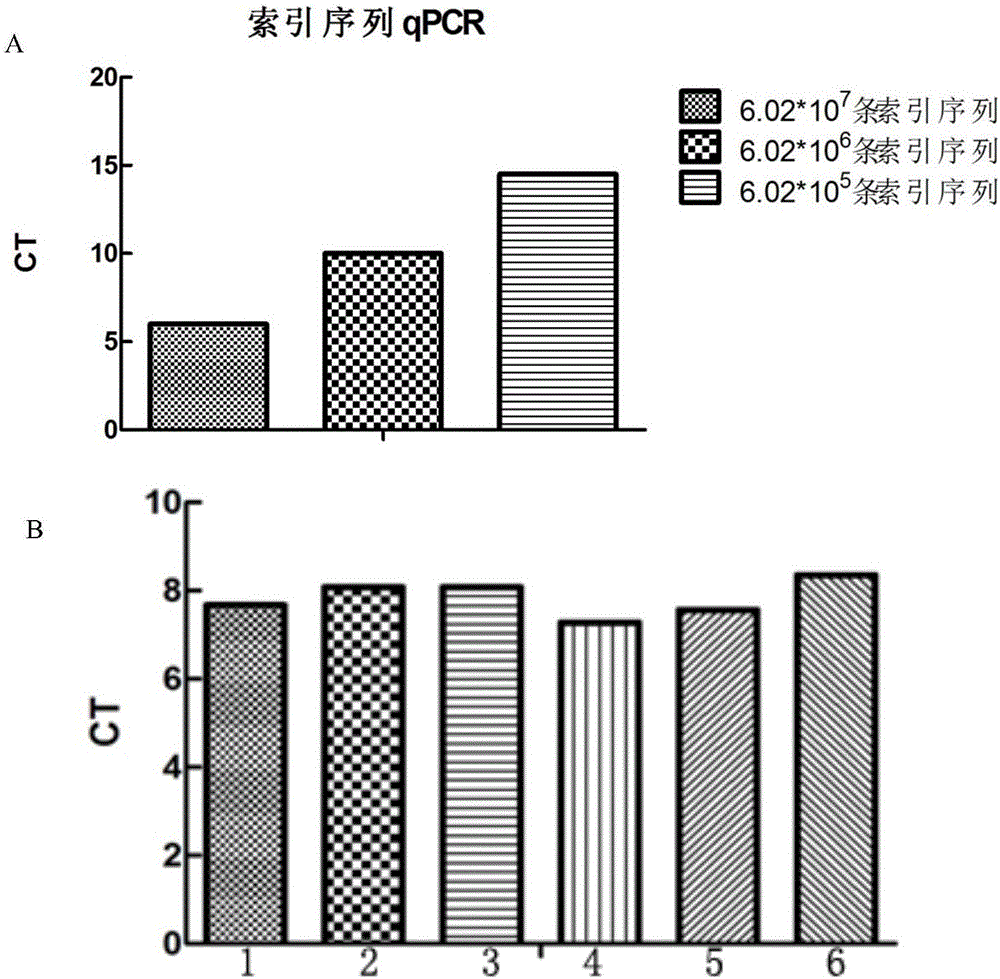
[0118] After the gene sequencing library was constructed, it was sent to Novogene for sequencing, using the HiSeq 4000PE125 sequencing strategy. Construction of quadruple CD34 using Microwell-seq high-throughput sequencing platform + The library is used as a duplicate, and the gene expression profile is obtained after splitting, screening and comparing. By importing this matrix file into R language analysis, the matrix data can be converted into a visualized graph. Figure 7 CD34 + Cellular gene expression difference heat map, it can be known that human CD34 + Cells can be divided into 7 subpopulations a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com