Nucleic acid for detecting A1166C polymorphic site of AGTR1 gene, kit and method
An A1166C, polymorphic site technology, applied in the field of nucleic acid and kits of A1166C polymorphic sites, can solve the problems of inability to meet the requirements of rapid clinical evaluation, unsuitable mutation site detection, restricted development and application, etc. , to avoid the problem of non-specific amplification, the detection results are reliable, and the detection effect is good.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Embodiment 1 primer, probe, verification template design
[0057] The selected ARMS primers of the present invention are designed for the A1166C SNP site. The selected ARMS primers are selected from a set of primer combinations that artificially add mismatched mutation sites at the 3' end and its vicinity. By screening primers that can distinguish single site alleles, a high degree of specificity.
[0058] After experimental verification, the specific detection primers for detecting the A1166C polymorphism site of the AGTR1 gene were finally determined to include wild-type upstream primers (AGRT1-FW), mutant upstream primers (AGRT1-FM), and common downstream primers (AGTR1-R). And public detection probe (AGTR1-P), nucleotide sequence is as follows:
[0059] AGRT1-FW (SEQ ID No. 1):
[0060] 5'-GCAGCACTTCACTACCAAATGAGTA-3';
[0061] AGTR1-FM (SEQ ID No. 2):
[0062] 5'-GCAGCACTTCACTACCAAATGAGTC-3';
[0063] AGTR1-R (SEQ ID No. 3):
[0064] 5'-AAAATGTGGCTTTGCTTTGTC...
Embodiment 2
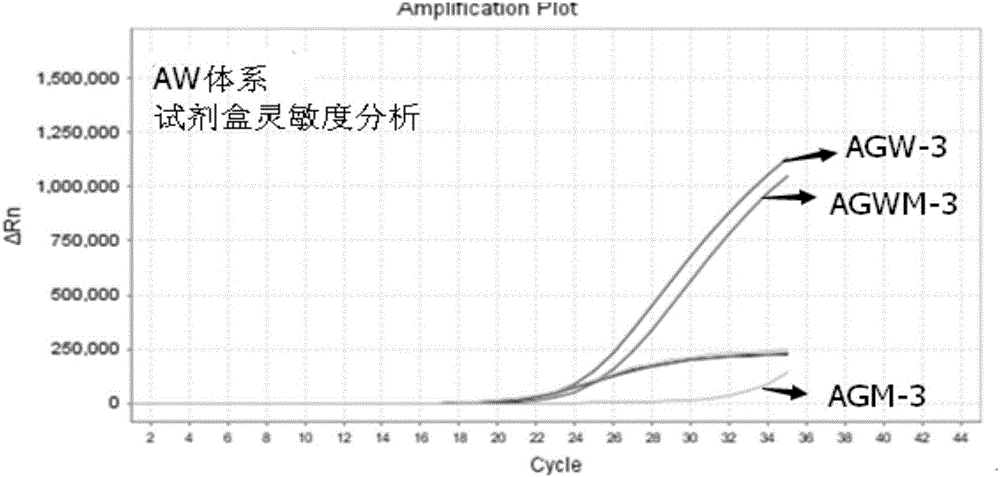
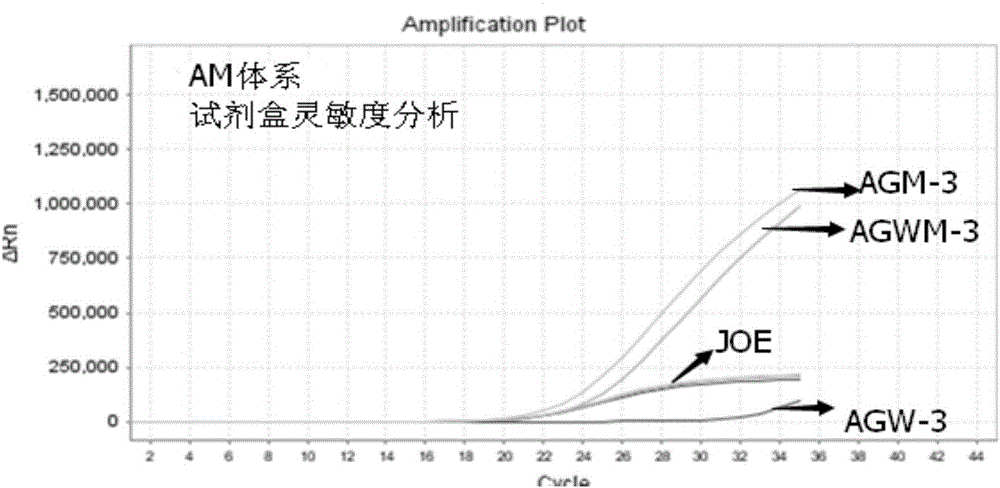
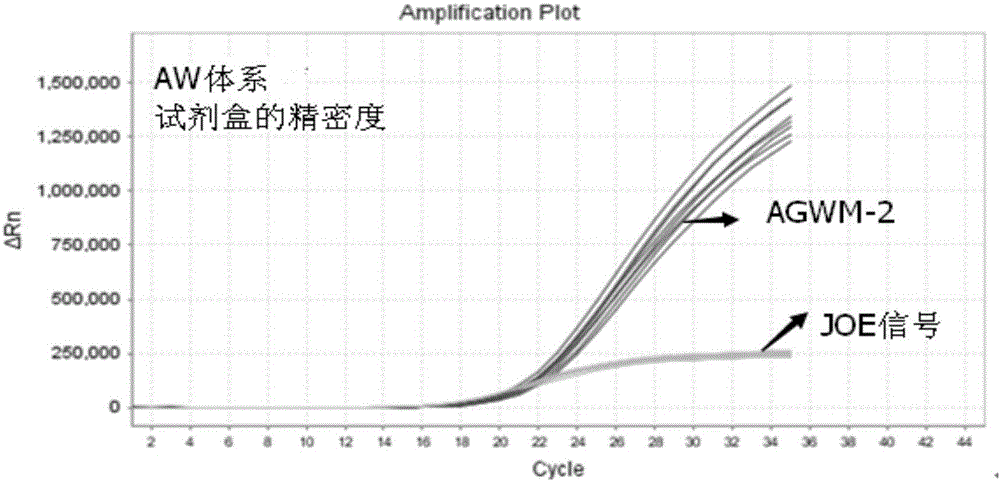
[0079] Example 2 The method for detecting the A1166C polymorphic site of the AGTR1 gene by real-time fluorescent PCR
[0080] The wild-type upstream primer, mutant upstream primer, public downstream primer and public detection probe designed according to Example 1 are synthesized, and a fluorophore can be connected to the 5' end of the public detection probe, and a quencher can be connected to the 3' end. Group, wherein the fluorescent group is any one of FAM, JOE, CY3, HEX, and the quenching group is any one of MGB, BHQ1, TAMRA, BHQ2.
[0081] Apply the wild-type primer pair, mutant-type primer pair and public detection probe designed by the present invention to the DNA template of the detection sample, and perform real-time fluorescent PCR amplification detection of the A1166C polymorphic site of the AGTR1 gene, using a wild-type reaction system and mutant reaction system, both use 40 μL reaction system as shown in Table 1;
[0082] Table 1 PCR reaction solution configurati...
Embodiment 3
[0090] Example 3 Kit for detecting the A1166C polymorphic site of the AGTR1 gene
[0091] The real-time fluorescent PCR kit for rapid detection of the A1166C polymorphic site of the AGTR1 gene includes the following components:
[0092] Wild-type upstream primer: the nucleotide sequence is shown in SEQ ID No.1;
[0093] Mutant upstream primer: the nucleotide sequence is shown in SEQ ID No.2;
[0094] Common downstream primers: the nucleotide sequence is shown in SEQ ID No.3
[0095] Public detection probe: the nucleotide sequence is shown in SEQ ID No.4, the 5' end of the public detection probe is connected to a fluorescent group, and the 3' end is connected to a quenching group;
[0096] Wild-type positive control: containing the nucleotide sequence shown in SEQ ID No.8;
[0097] Mutant positive control: containing the nucleotide sequence shown in SEQ ID No.9.
[0098] In order to avoid missed detection and wrong detection, it also includes: quality control primer pairs, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com