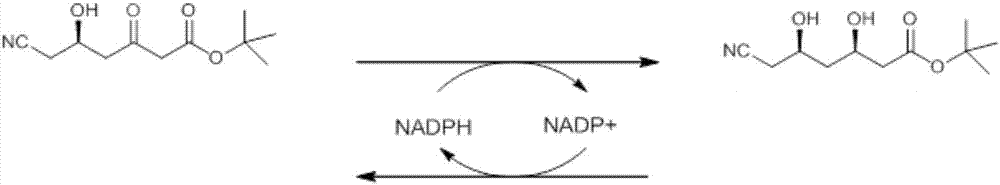
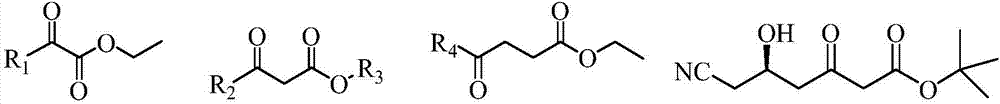
Carbonyl reductase, mutant and application of carbonyl reductase to preparation of statin synthetic intermediate
A technology of carbonyl reductase and reductase, which is applied in the direction of oxidoreductase, application, genetic engineering, etc., can solve the problems of increased cost, low catalytic activity, poor substrate tolerance, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The gene cloning of embodiment 1 carbonyl reductase LbCR
[0070] According to the open reading frame of carbonyl reductase LbCR, design upstream and downstream primers as follows:
[0071] The upstream primer, whose sequence is shown in SEQ ID No.3, specifically:
[0072] 5'-CGC GGATCC ATGACAGATCGTTTGAAAGATAAAGTGGCA-3';
[0073] The downstream primer, whose sequence is shown in SEQ ID No.4, is specifically:
[0074] 5'-CCC AAGCTT TTAAGCGCGTTGACCACCGTCAACCGTA-3';
[0075] Wherein, the underlined part of the upstream primer is the BamHI restriction site, and the underlined part of the downstream primer is the HindIII restriction site.
[0076] The genomic DNA of Lactobacillus brevis CGMCC 1.3258 was used as a template for PCR amplification. PCR system: 2×Taq PCR MasterMix 25μl, upstream primer and downstream primer (10ng / μl) each 2.5μl, genomic DNA (100ng / μl) 1μl and ddH 2 O 19 μl. The PCR amplification program was as follows: 32 cycles of denaturation at 95°C...
Embodiment 2
[0077] Example 2 Preparation of Carbonyl Reductase LbCR Recombinant Expression Plasmid and Recombinant Expression Transformant
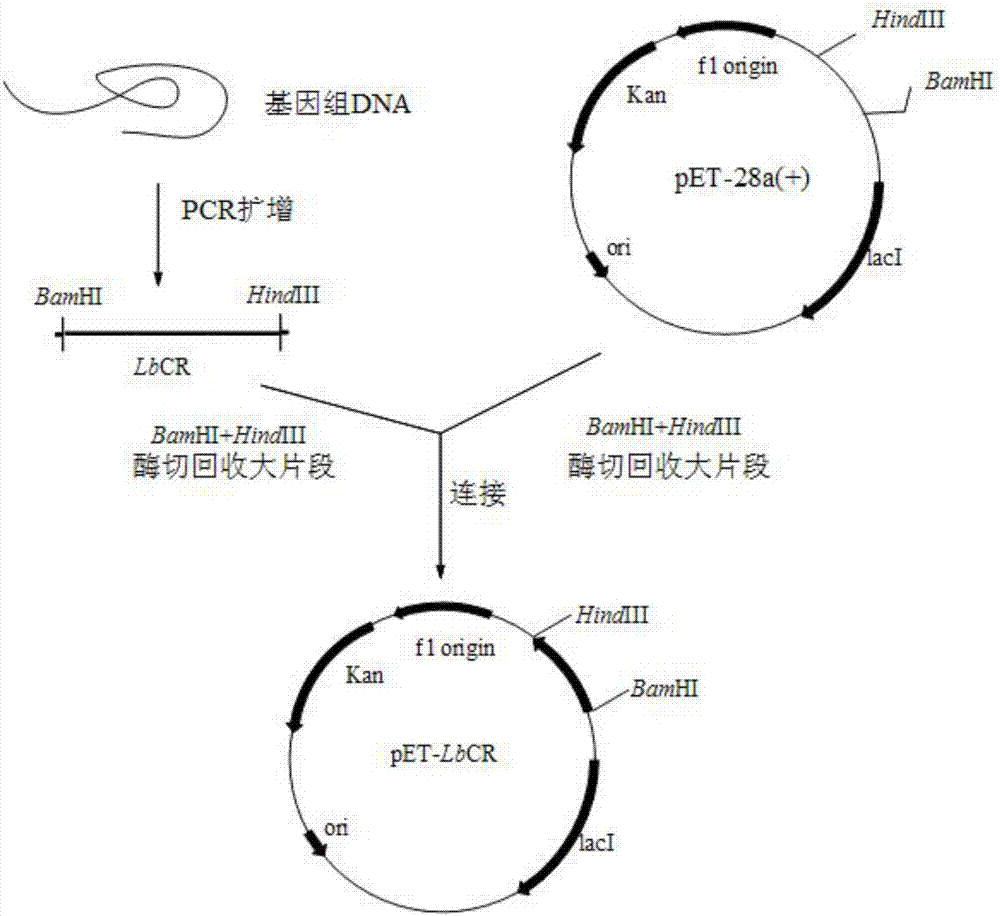
[0078] The carbonyl reductase target DNA fragment obtained by PCR amplification in Example 1 and pET 28a empty plasmid were double-digested overnight with restriction endonucleases BamHI and HindIII at the same time, then purified by agarose gel electrophoresis and recovered with a DNA kit. Ligate the recovered target fragment and the empty vector under the action of T4 DNA ligase at 4°C for 12 hours to obtain the recombinant plasmid pET28a-LbCR. The recombinant expression vector construction strategy of the gene is as follows: figure 1 shown.
[0079] Transform the resulting recombinant plasmid into E.coli DH5α, spread it on an LB medium plate containing 50 μg / ml kanamycin, and incubate it at 37°C for 8 hours. The grown colonies are verified by colony PCR and successfully amplified. Positive clones with a target band of about 750bp in length. Aft...
Embodiment 3
[0080] Example 3 Carbonyl reductase LbCR mutant construction
[0081] CASTing technology was used to construct the mutation library of carbonyl reductase LbCR: select non-conserved residues in the substrate binding pocket of LbCR for combinatorial saturation mutation, use degenerate codon NDT to design mutation primers, use pET28a-LbCR as a template, and use high-fidelity polymerization Enzyme PrimeSTAR for PCR. The PCR reaction conditions are as follows: to a PCR reaction system with a total volume of 20 μL, add 0.5–20 ng of template, 10 μL of 2×PrimeSTAR (Premix), 0.4 μL (10 μM) of each pair of mutant primers, and add sterilized distilled water to 20 μL. PCR reaction program: (1) denaturation at 98°C for 10 sec, (2) annealing at 55°C for 5 sec, (3) extension at 72°C for 60 sec, steps (1) to (3) were performed for 30 cycles in total, and the product was stored at 4°C. After the PCR product was verified by agarose gel electrophoresis analysis, the restriction enzyme DpnI was ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com