CrgA gene of Blakeslea trispora negative bacteria as well as cloning method and application of crgA gene
A kind of B. trispora and gene technology, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 3
[0030] Example 1 Extraction of B. trispora Genomic DNA.
[0031] Take the cultured B. trispora negative bacteria seed liquid, centrifuge to remove the supernatant, wash twice to obtain mycelia, then add an appropriate amount of liquid nitrogen to grind to white powder, take a small amount of DNA to extract, and refer to the fungal DNA extraction kit instructions.
Embodiment 2 3
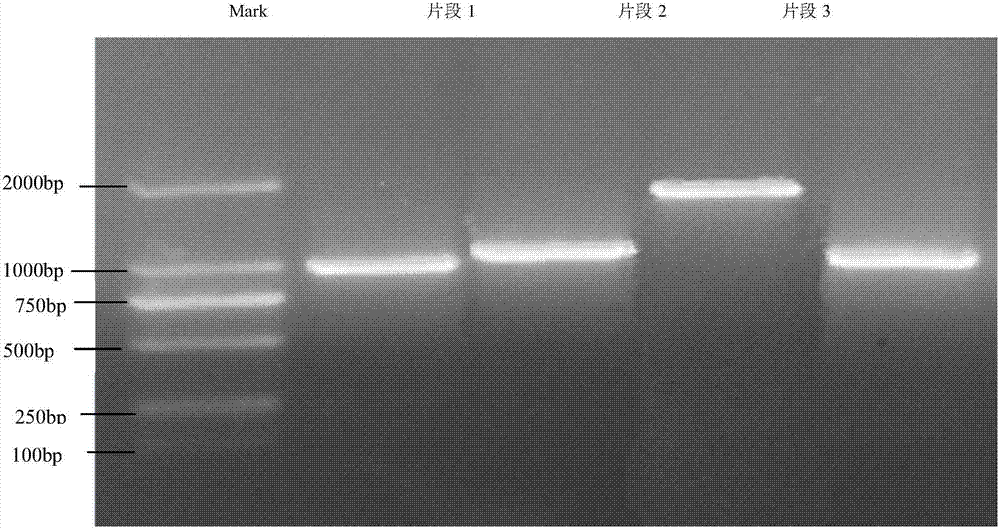
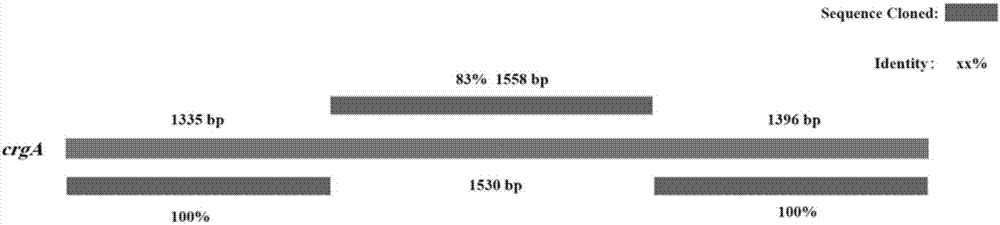
[0032] Example 2 Isolation and cloning of the crgA gene of B. trispora negative bacteria.
[0033] According to the sequence of the crgA gene of B. trispora, it was cloned in four segments and primers were designed respectively:
[0034] F1 (up): GGAAATTAAGCTATGCACCGCAGTATAGTC
[0035] F1 (down): AGGAAGGTTTGAACAGAAAACTCTTGTAGC
[0036] F2 (up): GGTAATGTATGTCGGTGTTGGTT
[0037] F2 (down): ACTCGGTTGAAGTGCGATTGTAT
[0038] F3 (up): AGTTAAAGCAATTCAAGCTTCGATTCTA
[0039] F3 (down): CTTTAAGAAATGCAACAAGTAGCAGGTG
[0040] F4 (up): ACAGACGACTGAAGAGATGATTGATGAACT
[0041] F4 (down): TATTTTCATATGGAACAAGATTTGTCTATA
[0042] The PCR reaction system is 50ul, including: Ex Taq enzyme 0.25uL, Ex Taq Buffer 5uL, dNTP Mix 4uL, template extracted from negative bacteria is less than 1ug, upstream / downstream primers (10uM) each 1uL, ddH 2 O up to 50ul; PCR reaction was carried out on the amplification instrument, and the reaction program was as follows: amplification conditions: pre-denatur...
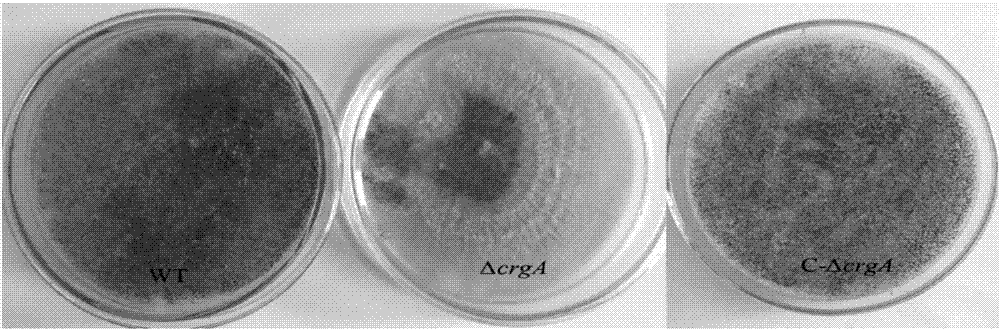
Embodiment 3
[0043] Example 3 takes the PMD-18T plasmid and Escherichia coli DH5ɑ as examples to illustrate the construction process of the prokaryotic expression system.
[0044] The product was identified by nucleic acid electrophoresis and gel-tapping recovery and purification. After purification and recovery, it was connected to the pMD-18T vector to construct a recombinant cloning plasmid, and then transformed into E. coli DH5α for amplification, and positive transformants were picked. LB liquid medium (AMPr) 37°C, 220rpm Cultivate for 10-12h, extract the plasmid, carry out double digestion and verification of the plasmid, and verify that the sequence is correct.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com