Method for producing glucaric acid by constructing recombinant saccharomyces cerevisiae and fermenting
A technology for recombining Saccharomyces cerevisiae and Saccharomyces cerevisiae, applied in the field of bioengineering, can solve problems such as MIOX activity decline and MIOX instability, and achieve high conversion rate, easy separation, and suitable for large-scale fermentation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Obtainment of BY4741opilΔSaccharomyces cerevisiae strain.
[0029] Using the pUG6 plasmid as a template, the primers PUG 6F and PUG 6R amplify the knockout frame (including the loxp-kan-loxp component), and the knockout frame has 50 bp bases in the upstream and downstream of the OPI1 gene as homology arms. Transform Saccharomyces cerevisiae BY4741 competent with lithium acetate chemical transformation method, spread the G418 resistance plate, and grow a single colony after 2-3 days. Use primers OPI-1 and OPI-4 to verify the correct bands by colony PCR, culture the correct strain in shake flasks to the logarithmic phase, and extract the genome for verification. This strain was used as a competent strain to transform the pSH65 plasmid, and galactose induced Cre to excise Kan. After verifying that the excision was correct, it was continuously passaged in YPD liquid medium to lose the plasmid pSH65, and the strain BY4741opi1Δ that knocked out the OPI1 gene was obt...
Embodiment 2
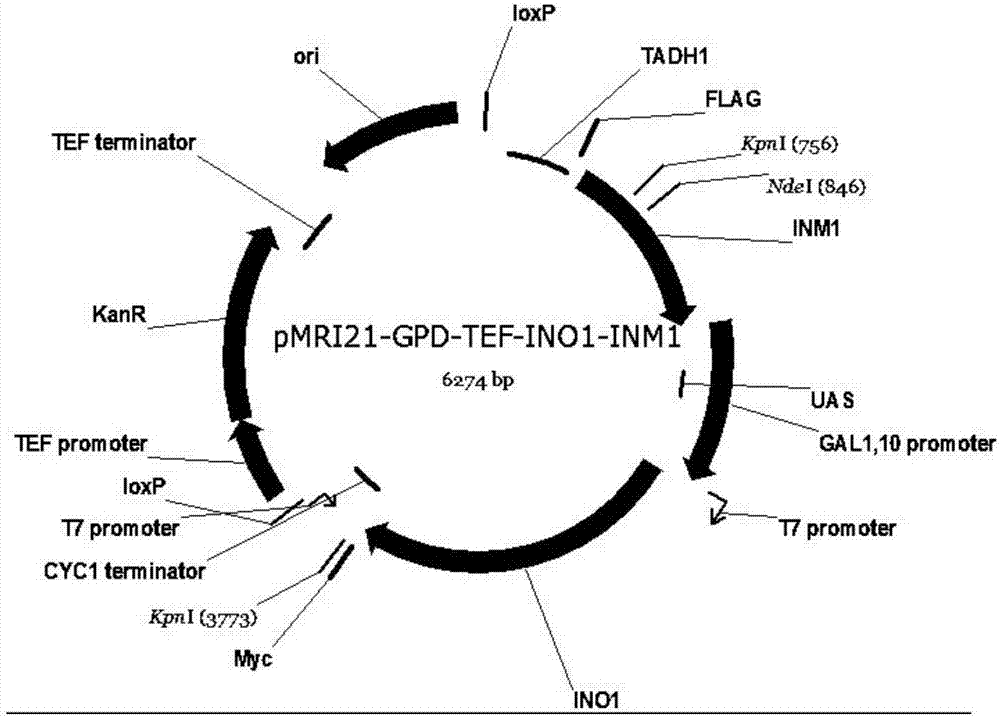
[0031] Example 2 Construction of recombinant strain BY4741opi1Δ-INO1-INM1
[0032] Select the constitutive plasmid pY26-GPD-TEF plasmid, use SmaI to single-digest the plasmid, use Gibson Assembly to assemble and construct pY26-INO1 plasmid; then use EcoRI to single-enzyme plasmid pY26-INO1, use Gibson Assembly to construct pY26-INO1- INM1 plasmid. Positive clones were verified using verification primers pY26F and pY26R, and the results were judged by the size of the electrophoresis band.
[0033] Plasmids Pmri-21 and pY26-INO1-INM1 were digested with BglII and SalI respectively, the Pmri21 plasmid and INO1-GPD-TEF-INM1 fragments were recovered by gel electrophoresis, and the corresponding cuts were connected with T4 ligase to obtain the recombinant plasmid Pmri-GPD- TEF-INO1-INM1, use BlpI single-digestion plasmid to linearize it, transform Saccharomyces cerevisiae BY4741opi1Δ, spread G418 resistance plate, and grow a single colony after 2-3 days. Colony PCR with primers INO...
Embodiment 3
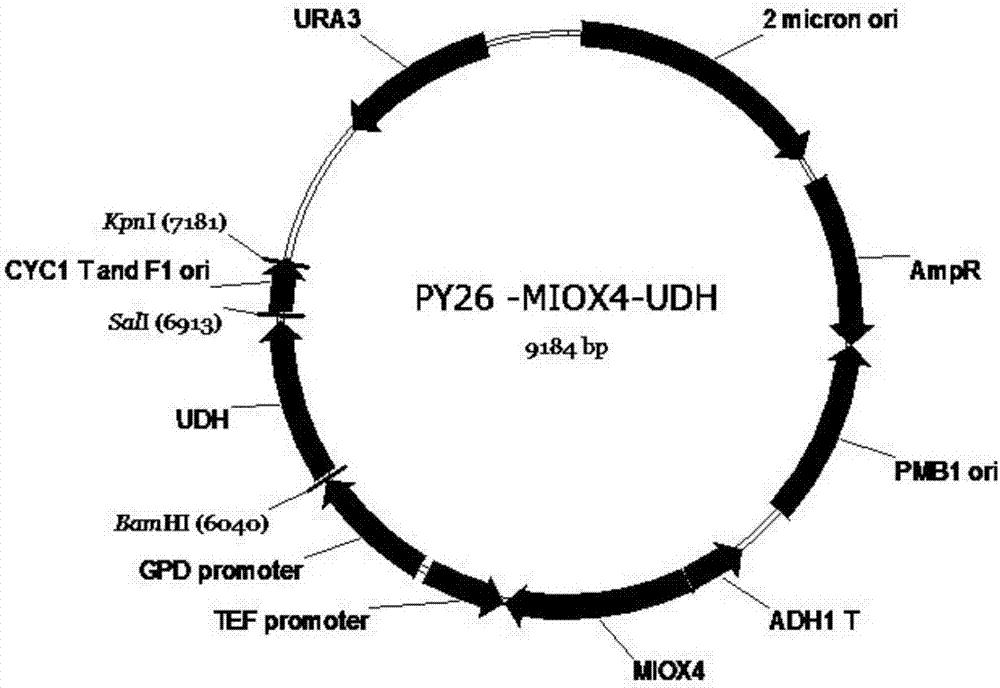
[0034] Example 3 Construction of recombinant bacterial group BY4741opi1Δ-INO1-INM / MIOX-UDH
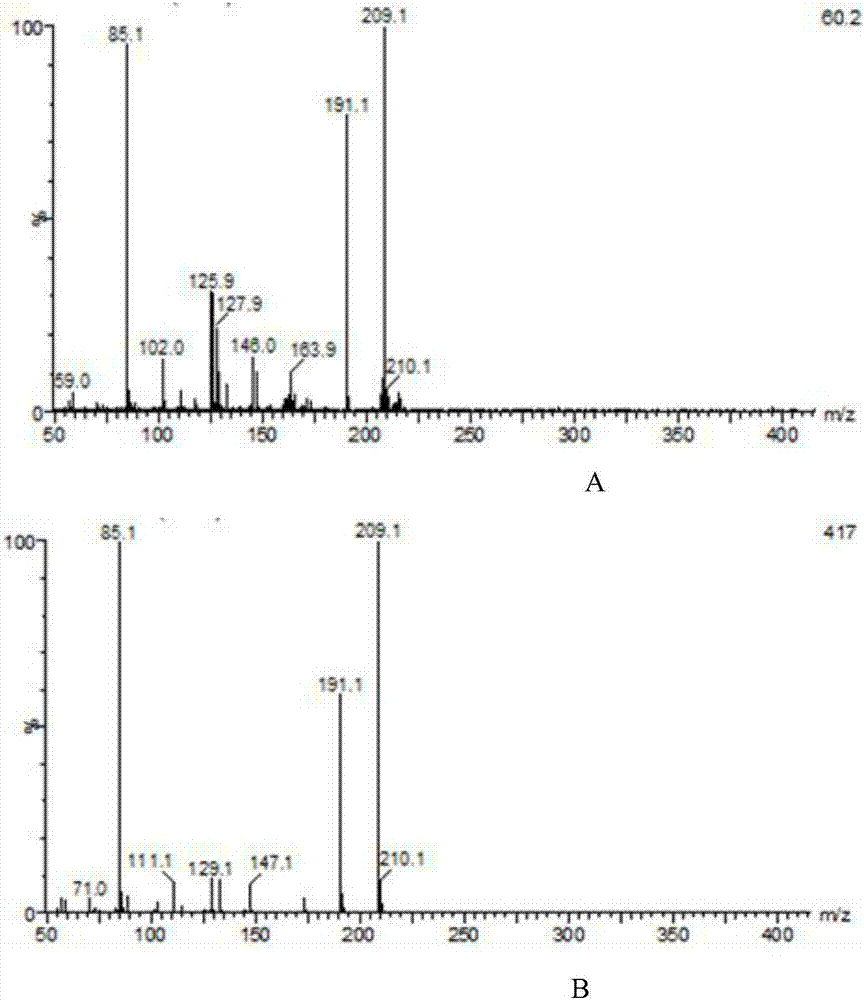
[0035] Fragment MIOX4 is from Arabidopsis thaliana, codon-optimized, as shown in SEQ ID NO:3, and inserted into pUC57 plasmid restriction site BglII / NotI to obtain pUC57-MIOX4. Fragment UDH is from Pseudomonas putida. After codon optimization, as shown in SEQ ID NO:4, it was inserted into EcoRI and SacII of pUC57-MIOX4 to obtain pUC57-MIOX4-UDH. Using pUC57-MIOX4-UDH as a template, using pY26-MIOX4F / pY26-MIOX4R and pY26-UDHF / pY26-UDHR as primers to amplify the MIOX4 and UDH genes respectively, purified by PCR product purification kit, and cut pY26 with SmaI -GPD-TEF plasmid, the fragments were recovered by the gel kit, and the pY26-UDH plasmid was assembled by GibsonAssembly; then the plasmid pY26-UDH plasmid was digested with EcoRI, and the pY26-UDH–MIOX4 plasmid was assembled by GibsonAssembly, and the primers were verified as pY26F and pY26R. Transform Saccharomyces cerevisiae BY4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com