A high-resolution conformation technique of genome-wide chromatin based on a small number of cells ehi-c 2.0
A chromatin and cell technology, applied in recombinant DNA technology, combinatorial chemistry, nucleotide library, etc., can solve the problems of poor data coverage and low interaction resolution.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Example 1. Preparation of eHi-C 2.0 sequencing library based on a small number of cells and its application in Hi-C sequencing
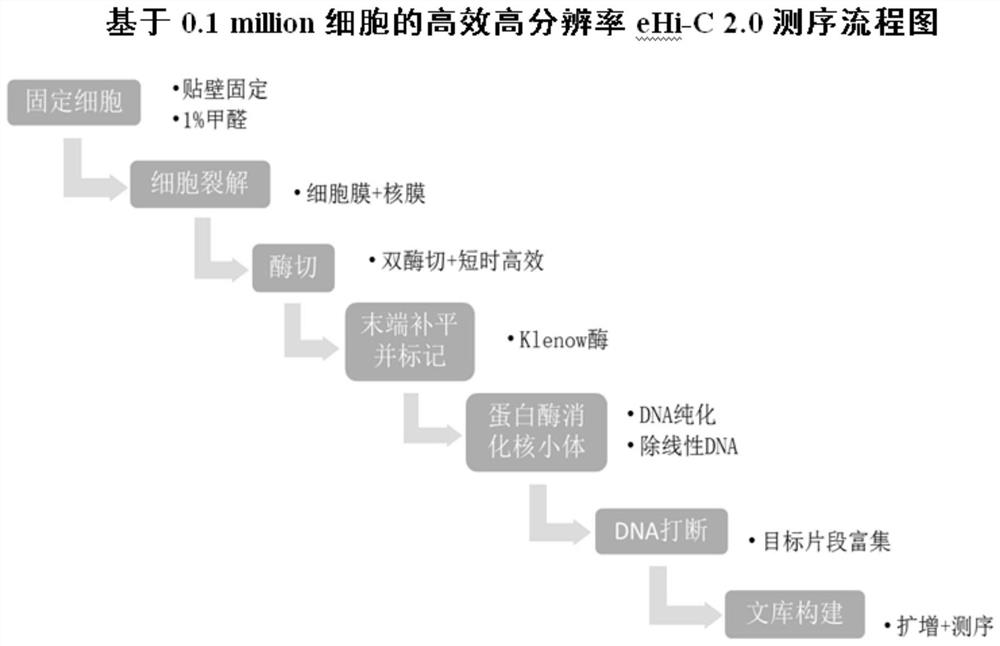
[0086] figure 1 It is a flowchart of high-efficiency and high-resolution eHi-C 2.0 sequencing based on 0.1 million cells.
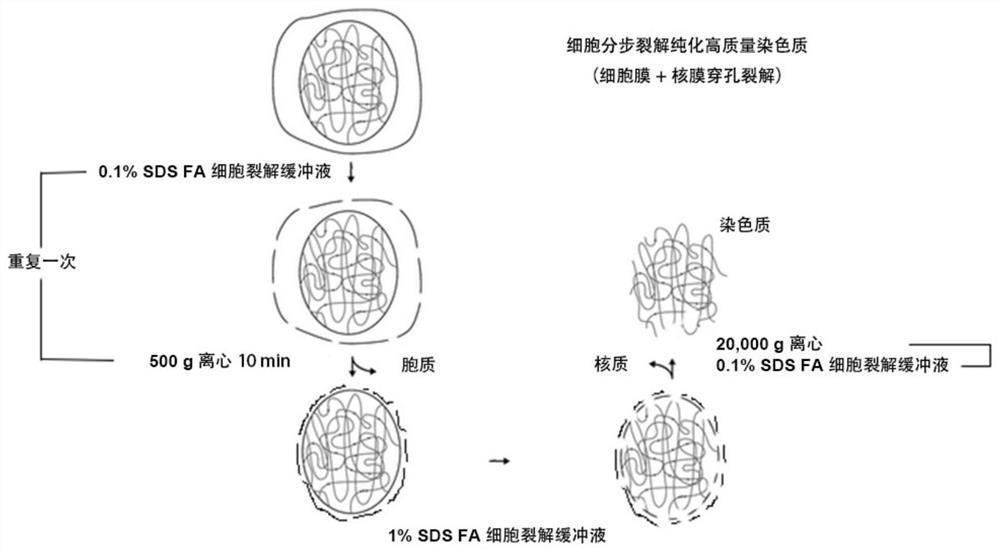
[0087] 1. Lysing cells to prepare chromatin ( figure 2 Optimize processing of major steps for stepwise lysis of cells. )
[0088] 1. Cell collection and cross-linking fixation
[0089] 1) When the cell confluency in the culture dish (10CM) reaches 80-90%, the C2C12 cells ( CRL-1772 TM ) about 6-8million (accurate result of the improved bovine Bauer cytometer);
[0090] 2) Add 37% formaldehyde aqueous solution (280 ul) in mass percentage to it to make the final concentration 1%, put the petri dish on a rotator and shake it at room temperature for 10 minutes (or incubate at 37° C. for 10 minutes);
[0091] 3) Add 2.5M glycine aqueous solution (0.89ml, so that the final concentration is 0.2M) to quench the formaldeh...
Embodiment 2
[0175] Embodiment 2, the restriction exonuclease combination selection test that eliminates linearized DNA
[0176] In Step 5 of Example 1, Lambda and Exonuclease I were used to eliminate the linearized DNA in the circular DNA mixture. The present invention did the following restriction exonuclease combination selection experiment when selecting the combination of these two enzymes for digestion, using PGL -4.23 Plasmids are tested as circular DNA mixtures:
[0177] 1. Acquisition of PGL-4.23 plasmid: Escherichia coli was used to transform, shake the bacteria, and extract the plasmid to obtain a plasmid with a concentration of 110ng / ul.
[0178] 2. Acquisition of linear pGL4.23-1: pGL4.23-1 is the pGL4.23 plasmid (Promega E8411) which was digested with BamHI. The system is as follows: plasmid 23.4ul; BamHI 5ul; 10×Cutsmart Buffer (NEB B7204s) 20ul; H 2 O151.6ul; Total 200ul. Digested at 37°C for 1 hour, purified with 1.2 times phenol chloroform after digestion, and the conce...
Embodiment 3
[0189] Example 3, Enzyme Combination Screening Test for Digesting Chromatin
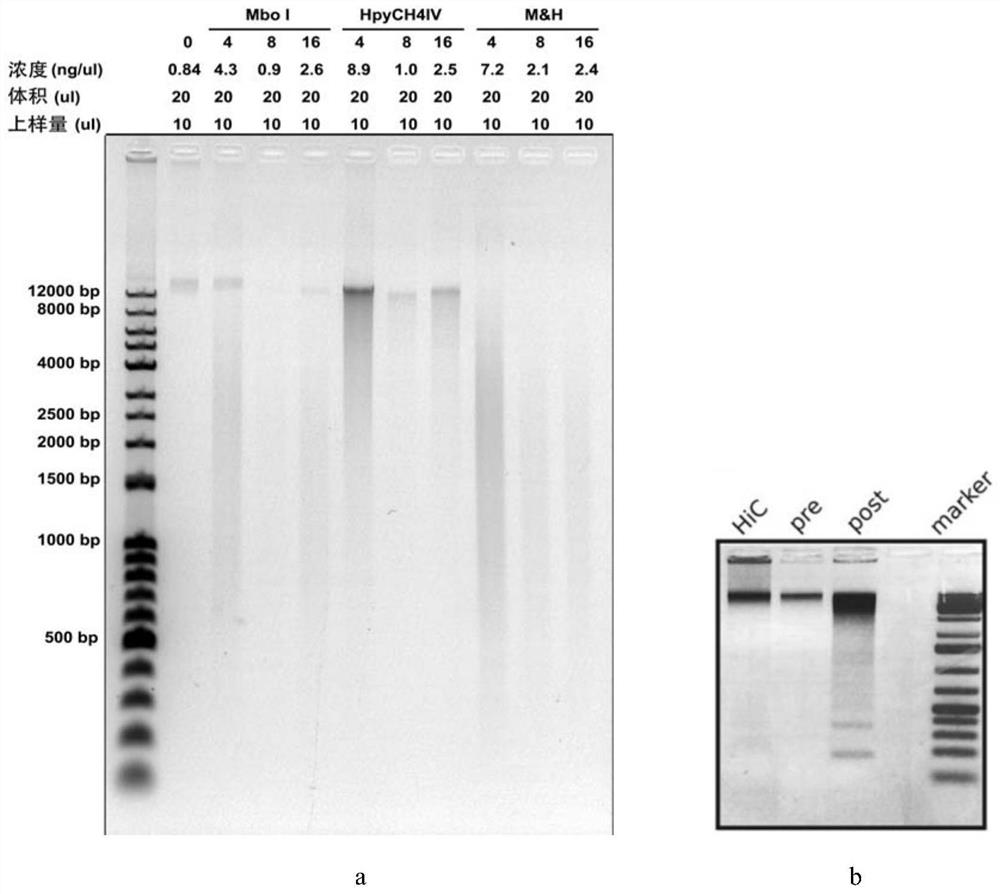
[0190] In step 2 of Example 1, MboI and HpyCH4IV were used to digest chromatin, and the present invention did the following enzyme combination selection experiment when selecting these two enzyme combinations for enzyme digestion:
[0191] 1. Take 6.6million cells / tube, add to 660ul, suck 10ul (that is, 0.1million) into ten 1.5EP tubes with Ruining white pipette tip, and carry out the first and second operations of Example 1, but in the second 3 ) step enzyme digestion time is divided into the following groups:
[0192] MboI single enzyme digestion group, add 2ul MboI (5U / ul, NEB R0147L);
[0193] HpyCH4IV single enzyme digestion group: add 2ul HpyCH4IV (10U / ul, NEB R0619L);
[0194] MboI and HpyCH4IV double restriction group (M&H): add 1ul MboI (5U / ul, NEB R0147L) and 1ul (10U / ul, NEB R0619L) of HpyCH4IV;
[0195] Each of the above groups was digested for 0h, 4h and 8h respectively (such as ima...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com