Porcine somatostatin gene editing site and application thereof
A technology of somatostatin and gene editing, applied in the fields of somatostatin, application, genetic engineering, etc., which can solve the problems of complex operation process, few applications, and high off-target rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Design and synthesis of sgRNA for specifically targeting SST gene in CRISPR / Cas9 specific knockout pig SST gene
[0031] 1. Design of sgRNA targeting porcine SST gene
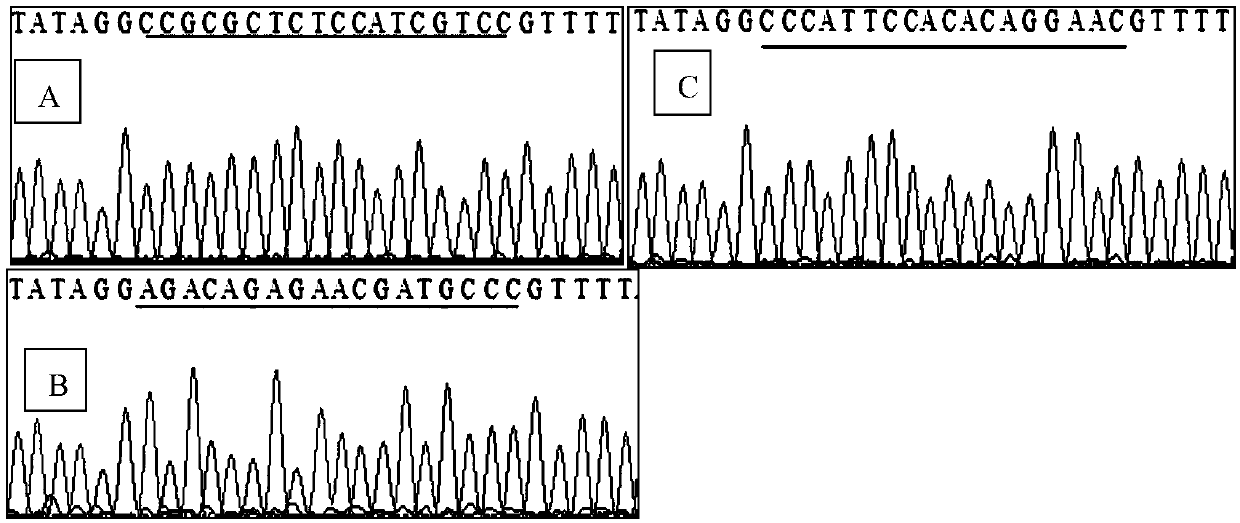
[0032] Using the website to design SST-sgRNA, the specific steps are: ① Log in to the GenBank website, search for the sequence of the porcine SST gene and download it, the nucleotide sequence of which is shown in SEQ ID NO:2. ② Open the target site prediction website (http: / / crispr.mit.edu / ), use the target site online design tool "CRISPR design tool", enter the gene sequence in the text box, select the species, and the gene sequence can be obtained For all target sites, several groups of target sites are selected after comprehensive consideration of various parameters. ③ The 20nt oligonucleotide sgRNA core sequence was designed according to the GG(N)19NGG sequence.
[0033] 2. Construction of sgRNA oligonucleotides
[0034] According to the selected sgRNA, add AAACACCG to the 5' end of the ...
Embodiment 2
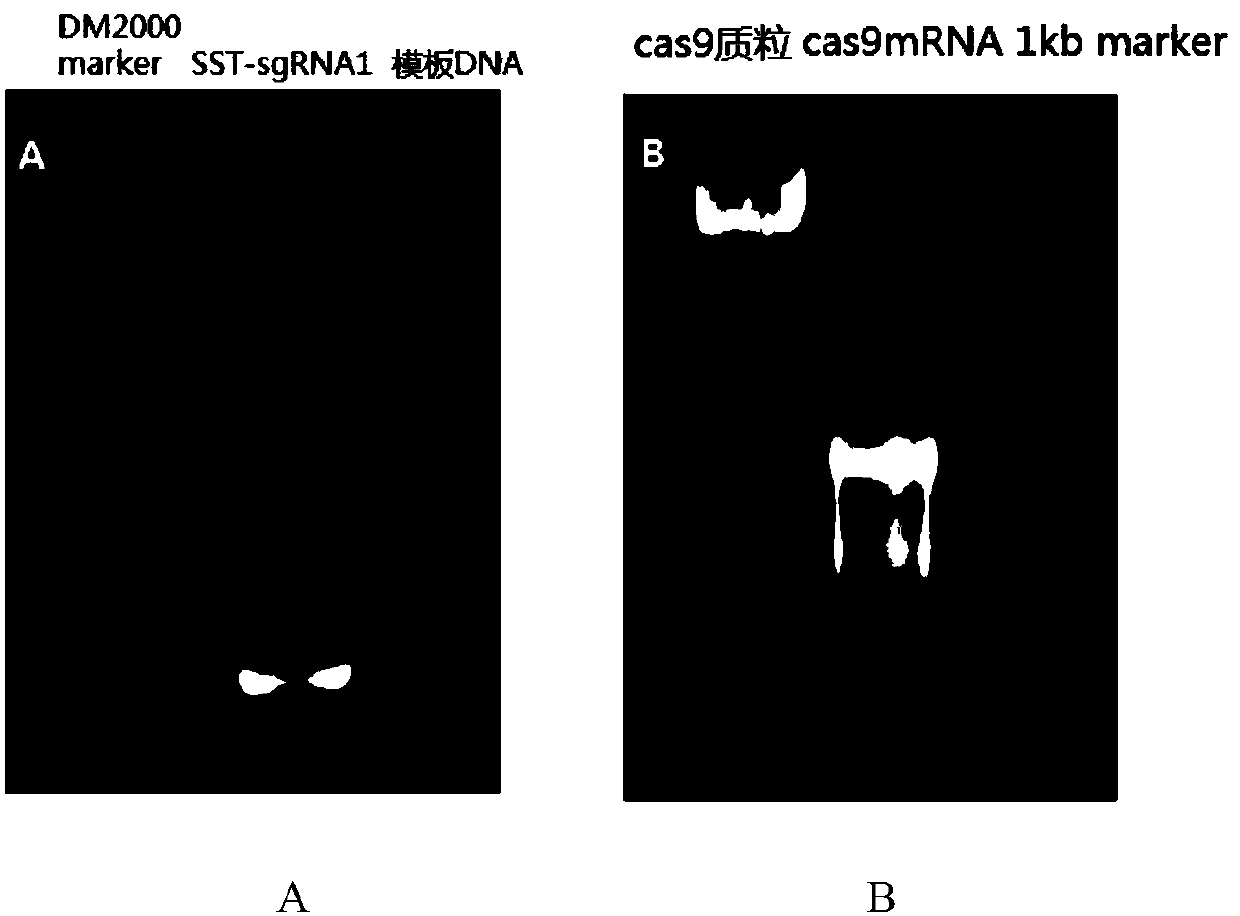
[0038] The preparation of embodiment 2sgRNA, Cas9mRNA
[0039] 1. Main reagents:
[0040] pmLM3613 (Addgene No.42251), BsaI, and PmeI enzymes were purchased from NEB Company; MEGA shortscript Kit (AM1354) sgRNA in vitro transcription kit, mMESSAGE mMACHINE Kit (AM1344) Cas9 in vitro transcription kit, Poly(A) Tailing Kit were purchased from Ambion Company; pDR274 (Addgene No.42250) plasmid was provided by Hubei Provincial Key Laboratory of Animal Embryo Engineering and Molecular Breeding; other commonly used reagents were domestic analytically pure; primer synthesis and DNA sequencing were completed by Shenzhen Huada Gene Company.
[0041] 2. Operation steps:
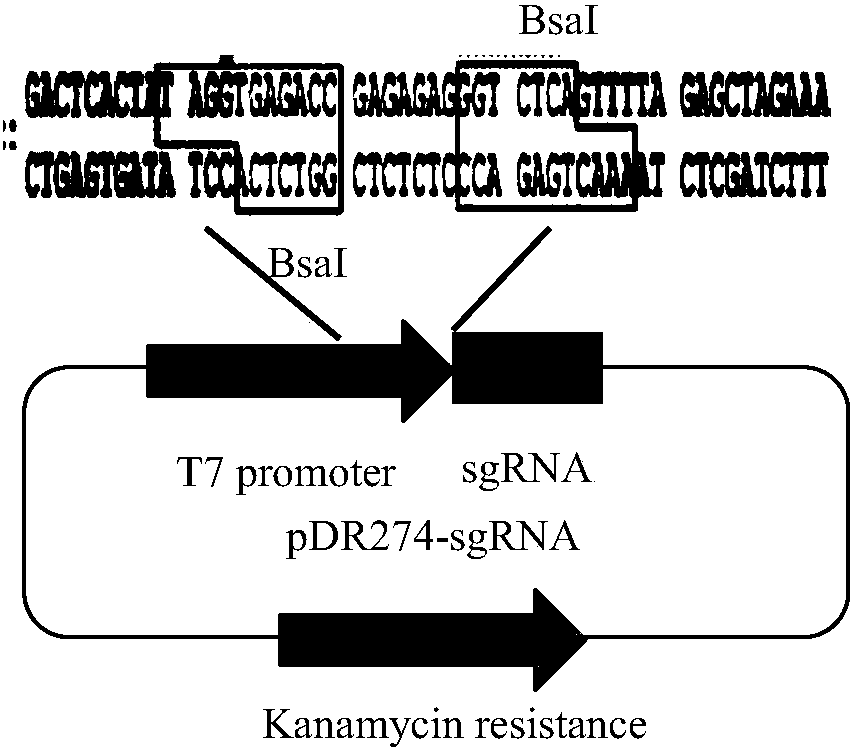
[0042] (1) In vitro transcription to obtain sgRNA:
[0043] 1) Construction of plasmid pDR274 expressing sgRNA (such as figure 2 shown).
[0044] ①Synthesis of double-strand oligonucleotide strands: sgRNA-g1 sites with high target activity add TAGG to the 5' end of the sense strand template, which is complementary to...
Embodiment 3
[0058] The collection of embodiment 3 oocytes and in vitro maturation culture
[0059] 1. Main solution preparation
[0060] (1) Preparation of oocyte maturation medium in vitro: the maturation medium is composed of improved TCM-199,
[0061] That is to add 3.05mmol / L glucose, 1mmol / L L-glutamine, 0.91mmol / L sodium pyruvate, 0.57mmol / L L-cysteine, 75ug / mL penicillin, 50ug / mL streptomycin sulfate. After mixing according to the above components, adjust the pH to 7.2-7.4, filter on the ultra-clean workbench, and store at 4°C for later use. 2 hours before the experiment, 10 IU / mL hCG, 10 IU / mL PMSG, and 10% pFF were added to the culture medium, and it was preheated in a constant temperature incubator for use.
[0062] (2) Preparation of embryo culture medium: composed of NCSU-23 basal medium + 4mg / mL BSA. NCSU-23 components are: 108.73mmol / L NaCl, 1.19mmol / L MgSO 4 , 25.07mmol / L NaHCO 3 , 4.78mmol / L KCl, 1.70mmol / L CaCl 2 , 5.55mmol / L glucose, 1.19mmol / L KH 2 PO 4 , 1.00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com