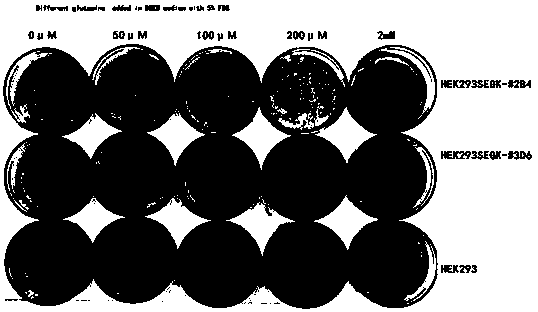
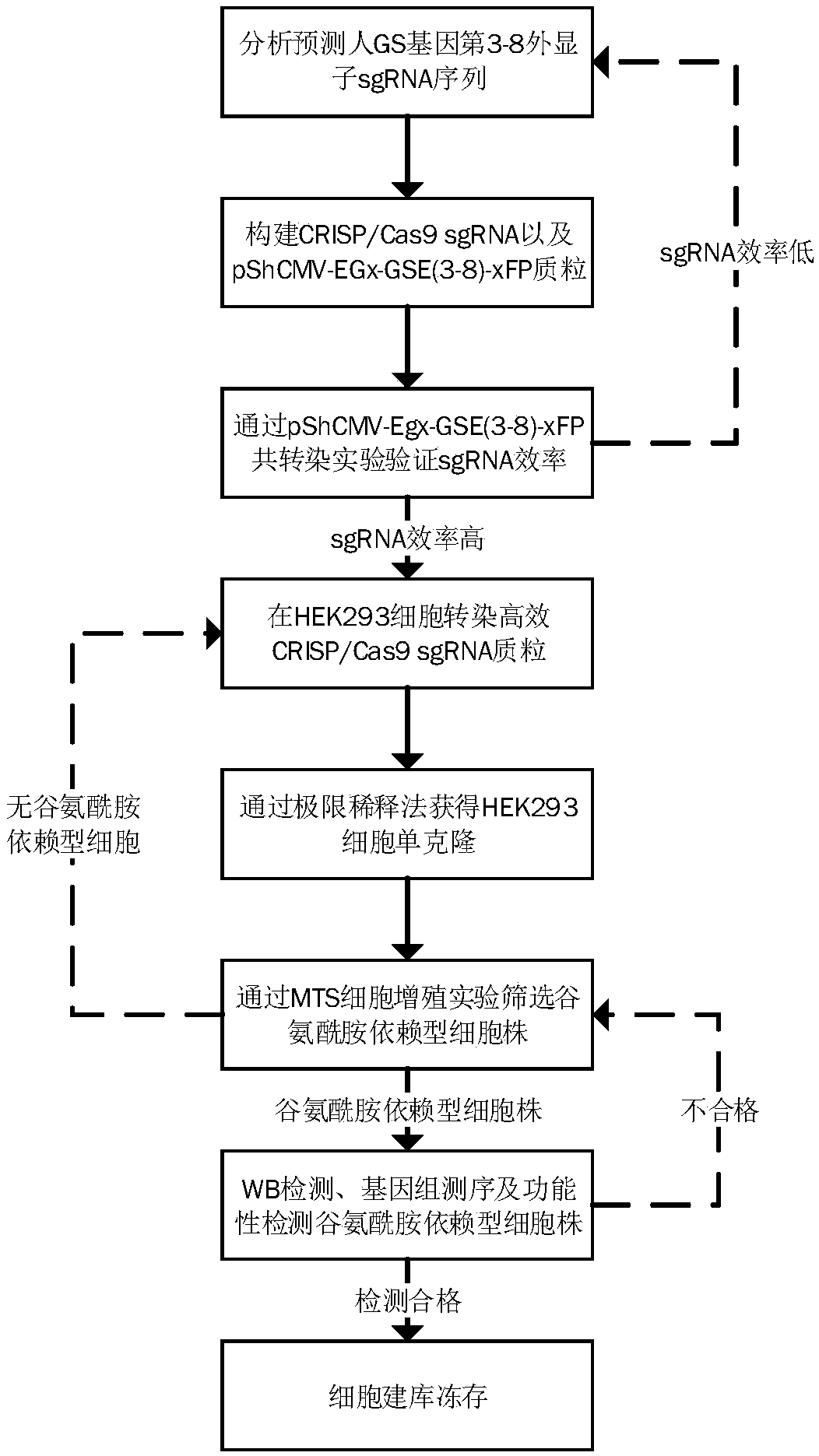
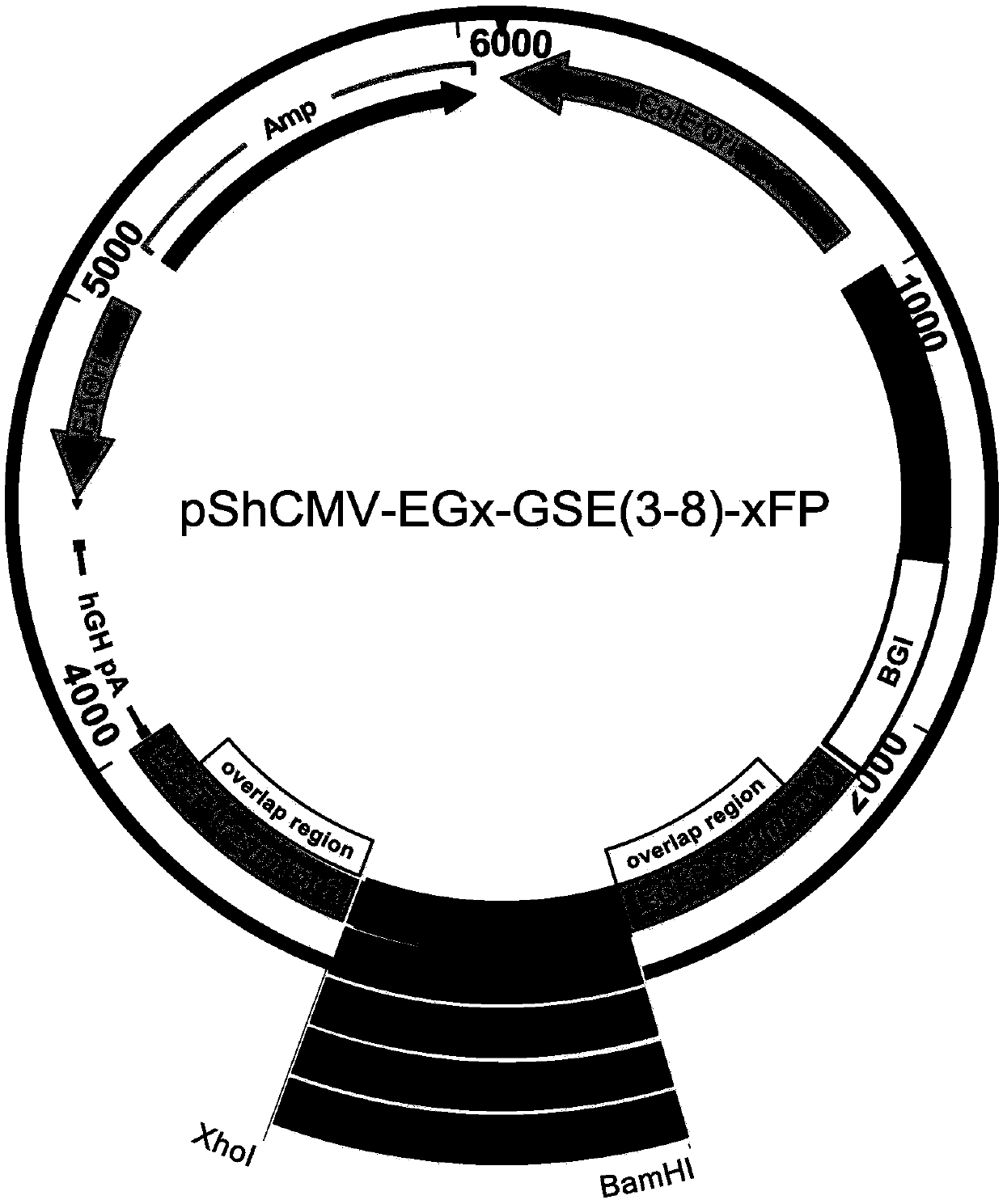
Method for screening glutamine synthetase-deficient HEK293 cell line
A glutamine and synthetase technology, applied in the biological field, can solve the problems of no screening ability, only 26% positive rate of cell screening, and inability to screen high gene copy number cell clones, etc., to simplify the culture process and have good clinical application prospects and commercial value, and the effect of improving the expression of recombinant protein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Example 1: Design GS gene sgRNA and construct CRISP / Cas9 plasmid
[0088] (1) Design GS gene sgRNA sequence
[0089] By comprehensively comparing the prediction results of various online tools, analyzing the number of mismatches, GC content, out-of-frame score and other conditions, in the third to eighth exon interval of the GS gene coding region ( SEQ ID NO.1) designed a total of 81 sgRNA sequences (see Table 1 for specific sequences).
[0090] Table 1 sgRNA sequence of GS gene coding region
[0091]
[0092]
[0093]
[0094] (2) Construction of pX330-GS-sgRNA (E3#01-E8#13) vector
[0095] Add BbsI cohesive end to the 5' end of the sgRNA sequence in Table 1, design the reverse complementary sequence, and synthesize corresponding primers (Table 2). The synthesized sgRNA forward and reverse complementary primers were dissolved in T4 polynucleotide kinase solution at a final concentration of 0.5 μM, and catalyzed by T4 polynucleotide kinase (T4 Polynucleotide K...
Embodiment 2
[0100] Example 2: Construction of pShCMV-EGx-GSE(3-8)-xFP plasmid
[0101] (1) Construction of pShCMV-EGx-MCS-xFP plasmid.
[0102] With plasmid pCMV(PacI)-MCS-IRES-EGFP (plasmid map as Figure 12 Shown) is the template PCR amplification enhanced green fluorescent protein (Enhanced Green Fluorescent Protein, EGFP) gene fragment. The primers for EGFPfrag1 were 5'acagATCGATgccaccATGGTGAGCAAGGGCGA G and 5'CTGggatccgaattcAGTGGTTGTCGGGCAGCAG; the primers for EGFPfrag2 were 5'TCACctcgagGCAAGCTGACCCTGAAGTTC and 5'CTACTGagatctTTACTTGTACAGCTCGTCCATG. The PCR reaction uses KAPAHiFiDNA Polymerase, the annealing temperature is 58°C, and the extension is 15s. The recovered EGFP1 fragment and pShCMV-MCS (MCS sequence is shown in SEQ ID NO.83, and the plasmid map is shown in Figure 13 (shown) the plasmid was digested with ClaI and BamHI, gel recovery and purification, T4 DNA ligase ligation, DH5α competent cell transformation and small amount of plasmid DNA preparation and identification...
Embodiment 3
[0107] Example 3: Rapid sgRNA efficiency detection
[0108] A total of 16 samples of pX330-GS-sgRNA (E3#01) to (E3#16) and pShCMV-EGx-GSE3-xFP plasmid; pX330-GS-sgRNA (E4#01) to (E4#10) and pShCMV- A total of 10 samples of EG x-GSE4-xFP plasmids (where E4#2-1 and E4#2-2 were co-transfected in equal mass ratios); pX330-GS-sgRNA (E5#01) to (E3#15) and pShCMV -15 samples of EGx-GSE5-xFP plasmid; pX330-GS-sgRNA(E6#01) to (E6#10) and pX330-GS-sgRNA(E7#01) to (E7#16) with pShCMV-EGx- A total of 26 samples of GSE(6&7)-xFP plasmid; a total of 13 samples of pX330-GS-sgRNA(E8#01) to (E8#13) and pShCMV-EGx-GSE8-xFP plasmid were co-transfected by calcium phosphate coprecipitation method HEK293 cells. The pX330 plasmid that does not contain the sgRNA sequence was mixed with pShCMV-EGx-GSE3-xFP, pShCMV-EGx-GSE4-xFP, pShCMV-EGx-GSE5-xFP, pShCMV-EGx-GSE(6&7)-xFP, pShCMV-EGx-GSE8 -xFP co-transfection with five plasmids served as a negative control. The experimental steps are as follows:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com