Application of enzyme FadD1 in pseudomonas aeruginosa for degrading DSF family signal molecules
A signal molecule, family technology, applied in the application, medical preparations containing active ingredients, hydrolase, etc., can solve the problems of no effect and unsatisfactory effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Obtaining of fadD1 gene sequence
[0033] BLAST alignment was carried out in the genome database of Pseudomonas aeruginosa PAO1 from the known amino acid sequence of Xanthomonas rpfB, and the sequence of fadD1 having 55% homology with rpfB was obtained, figure 1 It is the amino acid sequence alignment result of fadD1 and rpfB.
[0034] Using the Pseudomonas aeruginosa PAO1 genome as a template, design primers for PCR amplification, the amplification primers are:
[0035] fadD1-F: 5'-ATGATCGAAAACTTCTGGAAGGAC;
[0036] fadD1-R: 5'-TTACTTCTGGCCCGCTTTCTT,
[0037] After cloning analysis, the fadD1 gene and its sequence of Pseudomonas aeruginosa were obtained, as shown in SEQ ID NO: 1, the full length is 1689bp, the GC% content is 63.41%, the size of the protein encoded by it is 61.65kDa, and the amino acid sequence is shown in SEQ ID NO: 2 shown.
Embodiment 2
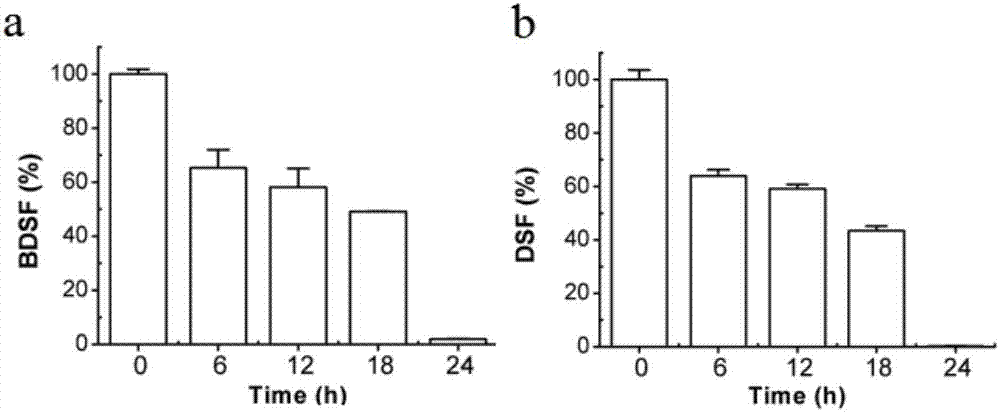
[0038] Example 2 FadD1 degrades signals BDSF and DSF in vitro
[0039] 1. Expression and purification of FadD1
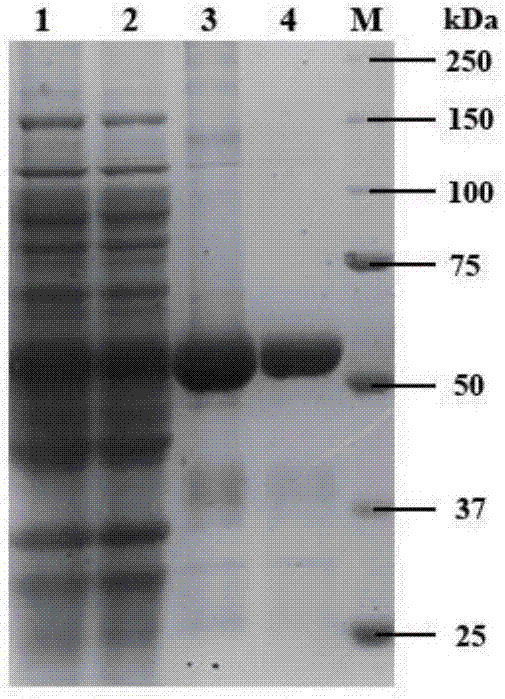
[0040] Construct the recombinant plasmid fadD1-pET-21a(+), transform the recombinant plasmid fadD1-pET-21a(+) into Ecoli.BL21(DE3), induce expression with IPTG, and analyze the expression product by SDS-PAGE, attached figure 2 Lane 1 and Lane 2 are supernatants of broken cells, and the size of the target protein FadD1 is 61.65kDa. The C-terminus of the target protein is connected with (His) 6 The tag was purified by NiSepharose 6 Fast Flow Beads by affinity chromatography.
[0041] 2. FadD1 degrades signal molecules BDSF and DSF in vitro
[0042] FadD1 degradation BDSF reaction system is: 5μM FadD1, 200μM BDSF, 30μL TME Buffer, 0.01MPBS Buffer to make up to 200μL, placed at 37°C for 6h, 12h, 18h, 24h respectively, then took out and boiled for 5min to terminate the reaction, and the reaction product was analyzed by LC-MS For detection, each treatment was repeate...
Embodiment 3
[0045] Example 3 Determination of the phenotypic effect of FadD1 on the regulation of the Burkholderia cepacia BDSF system
[0046] 1. Construction of Burkholder onion strain overexpressing gene fadD1
[0047] The overexpression plasmid fadD1-pBBRI-5 was constructed, and the plasmid was transformed into wild-type Burkholderia cepacia H111 (hereinafter referred to as H111), and a Burkholderia cepacia strain overexpressing gene fadD1 (abbreviated as H111 (fadD1 )).
[0048]2. Activation of Burkholderia cepacia
[0049] The Burkholderia cepacia wild-type strain H111, the Burkholderia cepacia mutant strain, the gene Bcam0581 related to BDSF biosynthesis in the H111 strain was knocked out (hereinafter referred to as mutant 0581), and the overexpressed gene fadD1 The strain H111 (fadD1) was activated on LB plates and cultured overnight in a 37°C incubator.
[0050] 3. Effect of FadD1 on strain biofilm
[0051] Get activated Burkholderia cepacia bacterial strain H111, mutant 0581...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com