A method for detection of nucleic acid structure based on surface-enhanced Raman spectroscopy
A surface-enhanced Raman and spectral detection technology, applied in Raman scattering, measuring devices, and material analysis through optical means, can solve problems such as high cost, difficulty, and impact, and achieve high reproducibility and reliability , the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
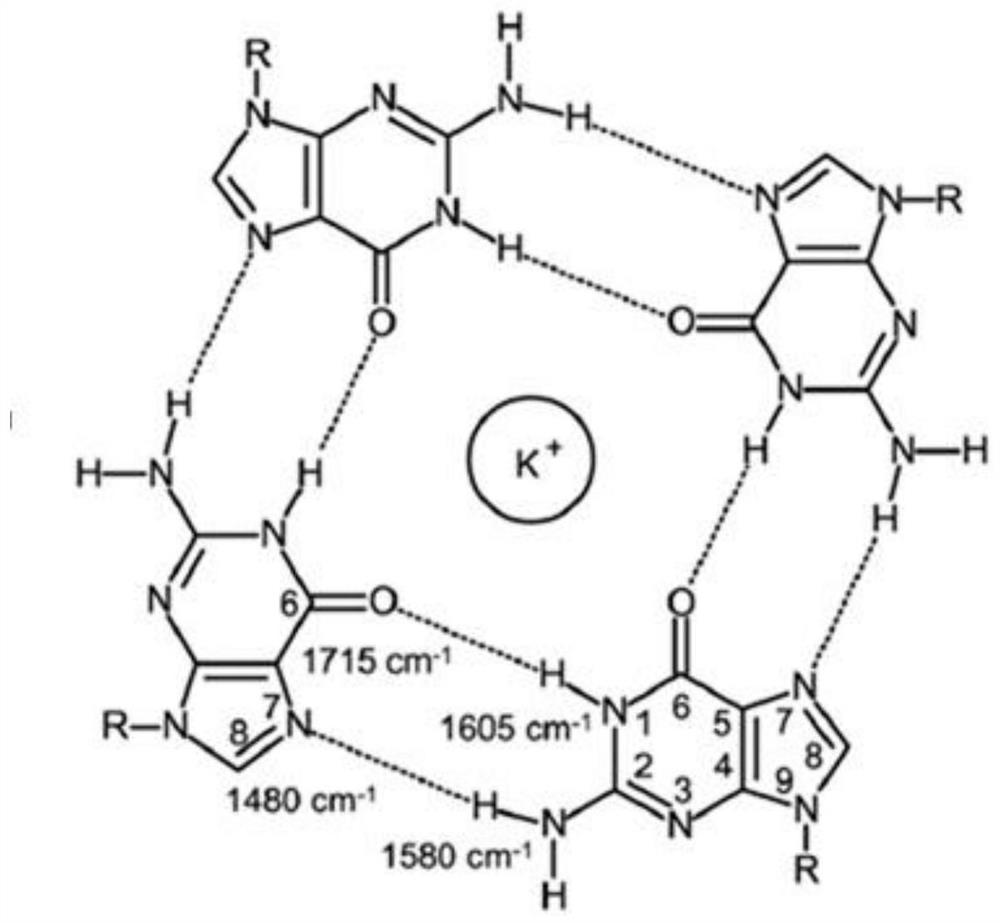
[0027] DNA generally exists in the form of double strands, but also forms triple-stranded structures and four-stranded structures. Such as figure 1 As shown, G-quadruplex is a kind of DNA four-strand structure, which is formed by the sequence rich in G (guanine) in Na+ or K+ solution, and each layer in the structure is between G and G The G-quartet is formed through Hoogsteen hydrogen bonds, and each base is a hydrogen bond donor and acceptor. This structure can consist of 4, 2 or 1 nucleotide chains. G-quadruplex is usually formed by stacking two or more G-quartets through planar bases. G-quadruplex is a ubiquitous DNA secondary structure in the human body. Detecting its structure includes the following steps:
[0028] (1) The preparation method of silver sol is with reference to classical lee method, and in the silver nitrate that 0.036g is dissolved in 200mL water, add the sodium citrate that 4ml mass concentration is 1%, the ultraviolet maximum absorption value of the ...
Embodiment 2
[0034] The difference between this embodiment and Embodiment 1 is that the pH of the buffer solution added in step (2) is 3.0.
Embodiment 3
[0036] The difference between this embodiment and Embodiment 1 is that the pH of the buffer solution added in step (2) is 5.0.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com