Baeyer-Villiger monooxygenase, mutant and application thereof in preparing long chain dicarboxylic acid
A technology of monooxygenase and dicarboxylic acid, applied in the direction of application, oxidoreductase, enzyme, etc., can solve the problems of shortening the synthetic route, poor regioselectivity, and low ratio of unconventional esters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
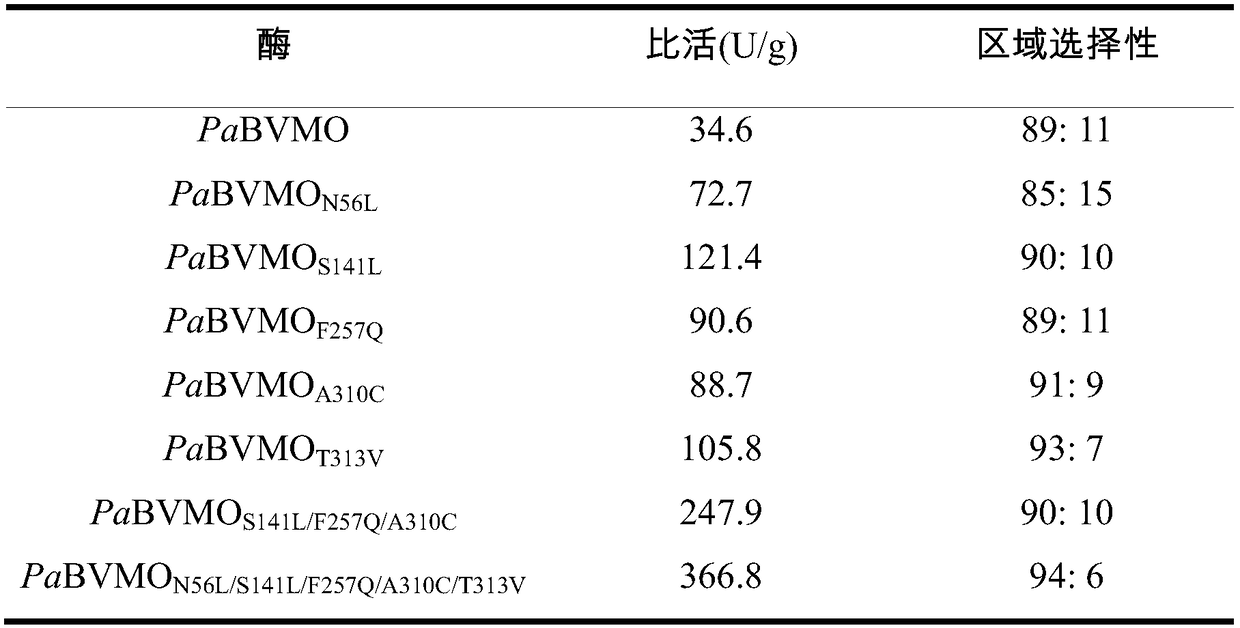
[0053] Example 1 Screening of "unconventional" regioselective Baeyer-Villiger monooxygenases
[0054] Using the Baeyer-Villiger monooxygenase PfBVMO with high "unconventional" region selectivity as the probe (sequence accession number AAC36351.2), through protein sequence alignment in NCBI, select a batch of similarities with the probe sequence For 20%-70% of the sequence, design primers for gene cloning. The target fragment amplified by PCR was digested and ligated into plasmid pET28a, transformed into E.coli DH5α, verified by colony PCR, and then sent to KingScript Biotechnology Co., Ltd. for sequence determination. The recombinant plasmids were extracted from the bacteria whose sequencing results were consistent with the target sequences, and the plasmids were transformed into E.coli BL21(DE3) again, and the target enzymes were overexpressed and regioselectively screened.
[0055] A 1 mL reaction was performed with recombinantly expressed whole cells using 1 mM 10-oxooctad...
Embodiment 2
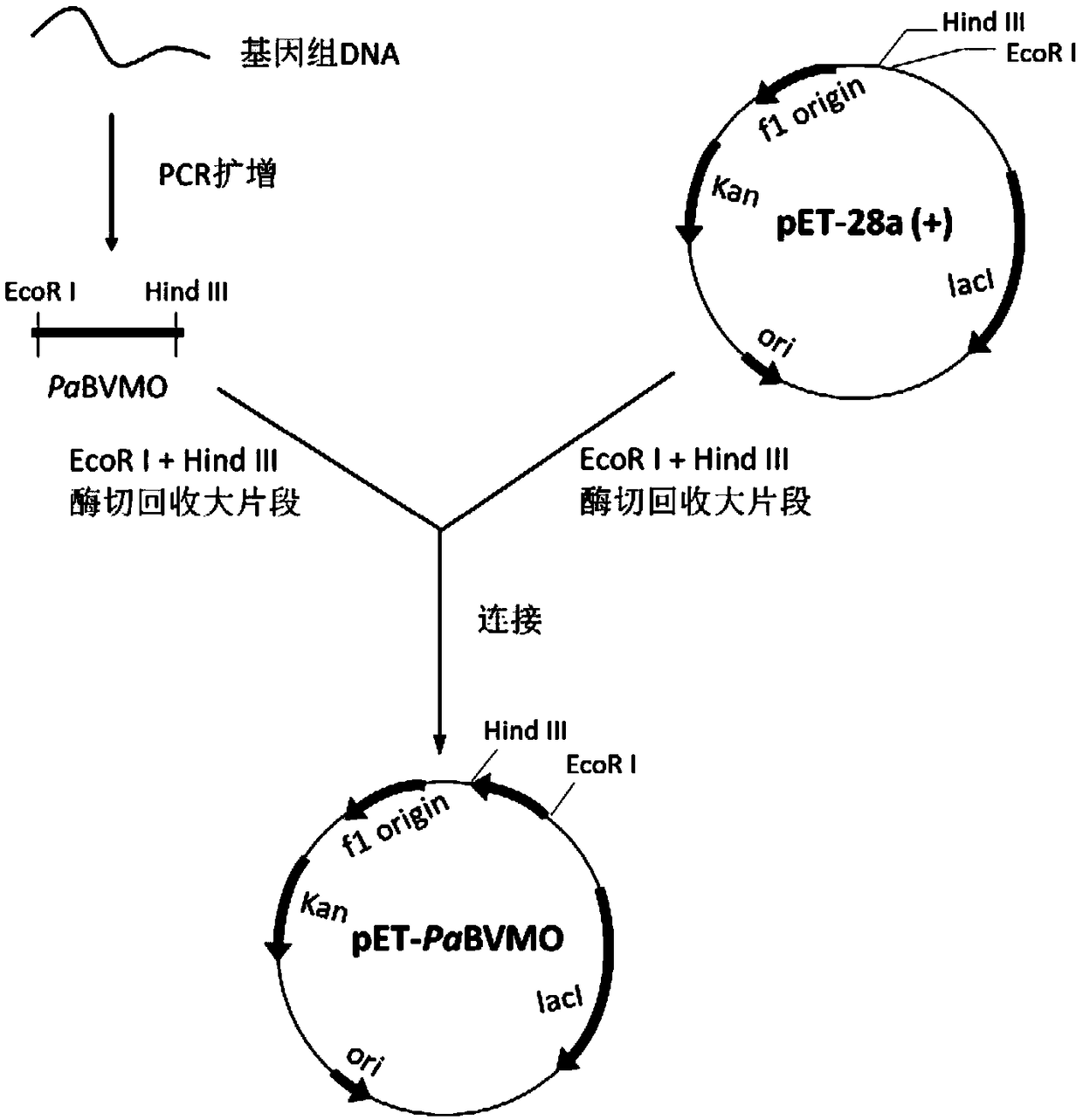
[0056] Cloning and recombinant expression transformant construction of embodiment 2 monooxygenase PaBVMO gene
[0057] According to the gene sequence (Genebank accession number: WP_003087250.1) recorded in Genebank in Pseudomonas aeruginosa and predicted to be Baeyer-Villiger monooxygenase, the PCR primers were designed as follows:
[0058] Upstream primer: CCG GAATTC ATGAGTACCCAACCCCACC;
[0059] Downstream primer: CCC AAGCTT TCATGCGGGTACCCCCTTC.
[0060] Wherein, the underlined part of the upstream primer is the restriction site of EcoR I, and the underlined part of the downstream primer is the restriction site of Hind III.
[0061] The genomic DNA of Pseudomonas aeruginosa (Pseudomonas aeruginosa) with the preservation number CGMCC1.9047 was used as a template for PCR amplification. The total volume of the PCR amplification system is 50 μL, specifically: 2x Taq PCR MasterMix 25 μL, upstream primer and downstream primer 1.5 μL (0.4 μmol / L), DNA template 2 μL (1 μg) and ...
Embodiment 3
[0062] Random mutation of embodiment 3 monooxygenase PaBVMO
[0063] Using error-prone PCR technology to introduce random nucleotide mutations into the PaBVMO gene, the primers used are as follows:
[0064] Upstream primer: CCGGAATTCATGAGTACCCAACCCCACC;
[0065] Downstream primer: CCCAAGCTTTCATGCGGGTACCCCTTC.
[0066] Wherein, the template is the recombinant plasmid of PaBVMO gene obtained as in Example 2.
[0067] The PCR reaction system (50μL) is: template 0.5-20ng, 10×rTag buffer 5μL, dNTP (2.0mM each) 5μL, MgSO 4 (25mM) 2μL, MnCl 2 (100 μM) 5 μL, a pair of mutant primers (20 μM) 1 μL each, 1 unit of rTaq enzyme, add ultrapure water to 50 μL.
[0068] The program of PCR amplification is: (1) Denaturation at 94°C for 3min; (2) Denaturation at 94°C for 10s, (3) Annealing at 60°C for 30s, (4) Extension at 68°C for 90s, steps (2)-(4) were carried out for 30 seconds in total. The final cycle was extended at 72°C for 10 min, and the product was stored at 4°C.
[0069]After ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com