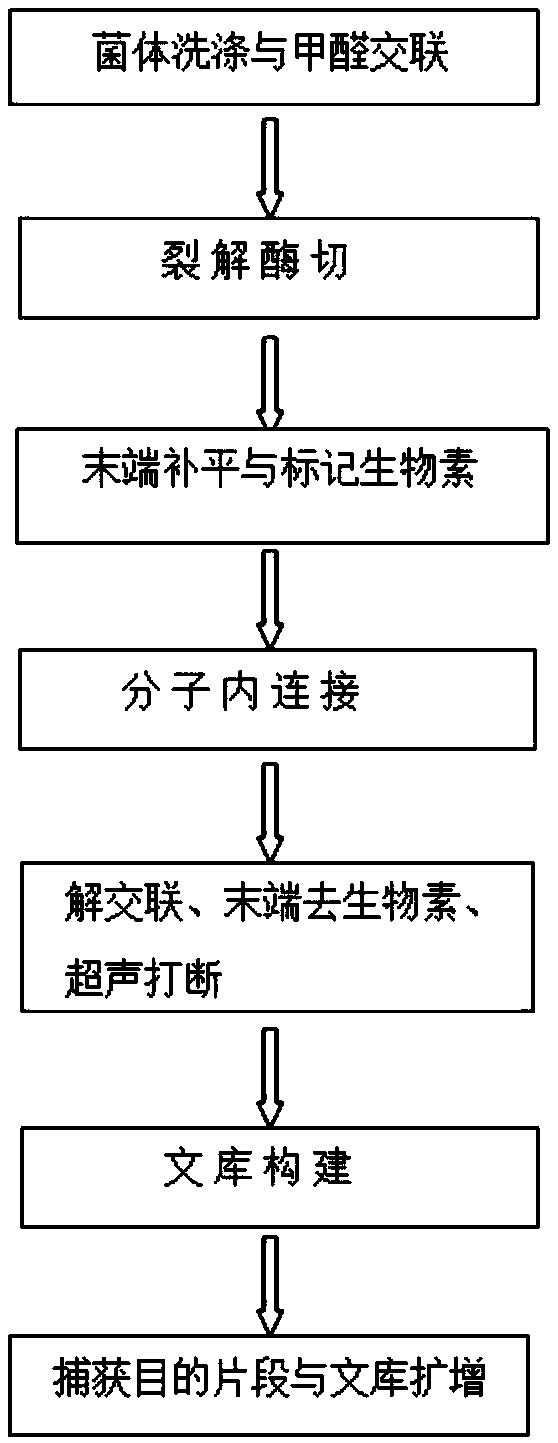
Hi-C high-throughput sequencing library construction method applicable to plants
A high-throughput, plant technology, applied in the field of molecular biology, can solve the problems of poor library construction effect of difficult-to-release cytoplasmic plant samples, and achieve the effects of improving integrity and cross-linking effect, shortening reaction time, and simplifying operation steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] In this example, healthy soybean leaves were taken as the research object. Fresh leaves were cross-linked with formaldehyde, cells were lysed to release chromatin, and then the chromatin was digested, biotin-labeled, intramolecularly connected, and ultrasonically interrupted, and finally the library was constructed. The specific experiment The process is as follows:
[0052] 1. Formaldehyde fixation
[0053] 1) Take 1 g of young soybean leaves (preferably without a waxy layer), cut them into pieces about 0.5 cm in size, put them into a 50 mL centrifuge tube, and add 35 mL of pre-cooled NIB lysate to the centrifuge tube (add 35 μL of mercapto ethanol and 35 μL of 100 mM PMSF) and 3 mL of about 37% formaldehyde solution (final concentration 2%), vacuumize on ice for 40 min;
[0054] 2) Add 7.5mL 2.0M glycine and treat under vacuum for 5min to terminate cross-linking;
[0055] 3) Remove the solution and wash 2-3 times with sterilized ultrapure water, and completely dry t...
Embodiment 2
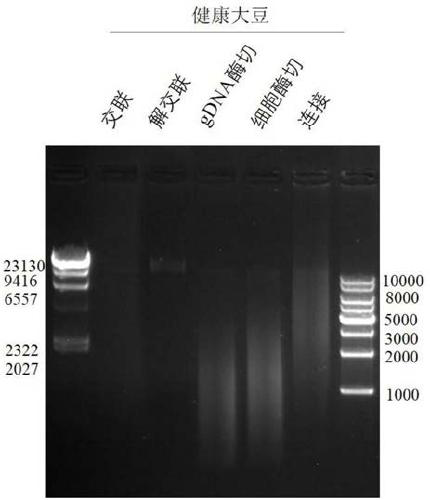
[0108] In this example, soybean leaves with diseases and insect pests were taken as the research object. The soybean leaves with disease and insect pests were cross-linked with formaldehyde, the cells were lysed to release chromatin, and then the chromatin was digested, biotin-labeled, intramolecularly linked, and ultrasonically interrupted, and finally the library was constructed. The experimental process is the same as in Example 1. Identify the genome integrity, enzyme digestion effect, and connection effect by agarose gel electrophoresis pattern. The agarose gel electrophoresis pattern is as follows: Figure 4 As shown in the picture, the complete genomic DNA bands of soybean leaves affected by diseases and insect pests are obviously without tailing; the enzymatic digestion effect is obvious, and the entire DNA fragment moves down; Proceed to the next experiment.
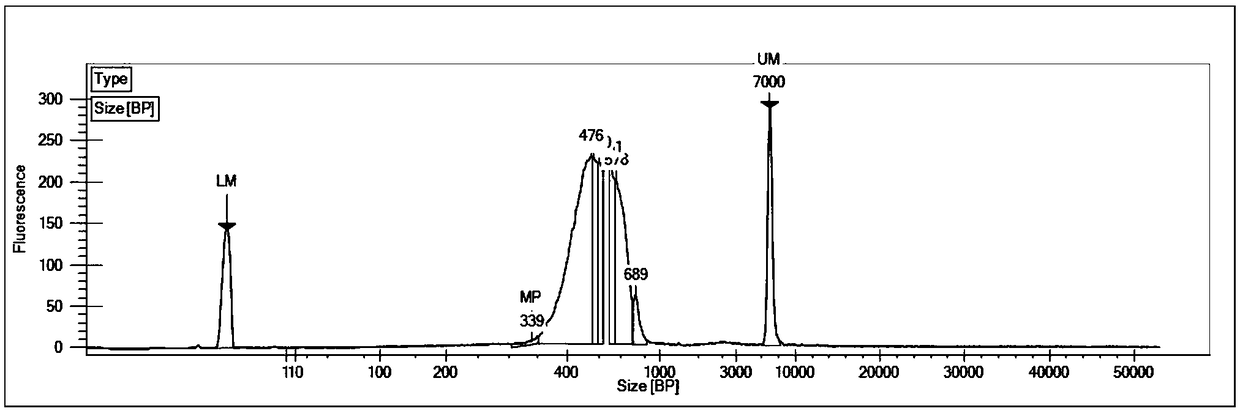
[0109] For the clean data obtained by quality control, use ICE3 software to iteratively compare the data and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com