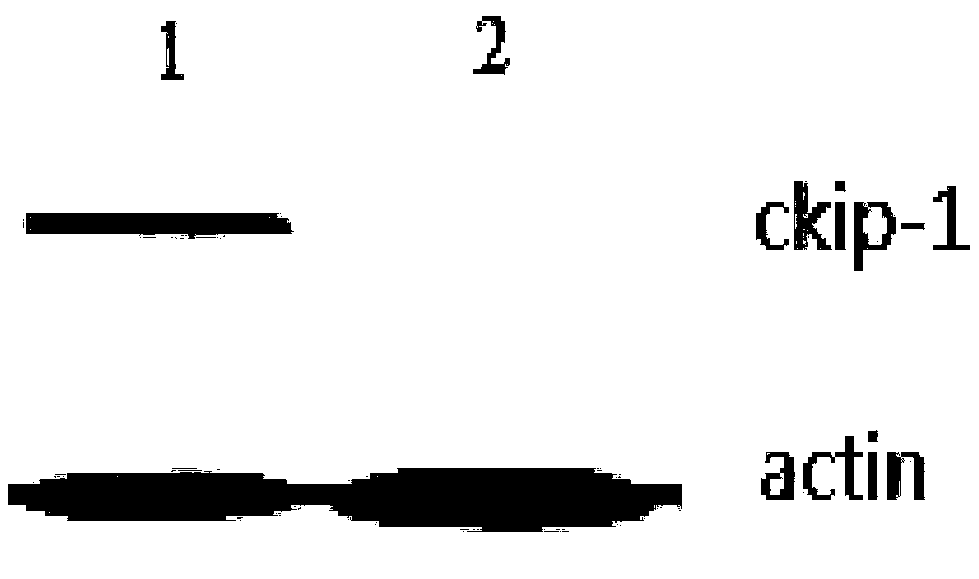CKIP-1 RNAi compound of macrophage targeted and preparation method and application thereof
A technology for targeting macrophages and complexes, applied in the field of CKIP-1 RNAi complex and its preparation, can solve the problems of high fatality rate and disability rate, lack of treatment methods, etc., to reduce brain edema and enhance transfection efficiency , the effect of increasing the uptake rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Preparation of embodiment 1 fusion peptide
[0041] The fusion peptide is formed by directly linking the carboxyl terminal of the V6 peptide, the amino terminal of the T9 peptide and the amino terminal of the V6 peptide in sequence, and the amino acid sequence is SEQ ID NO.3.
[0042] The fusion peptide was synthesized on an AB-431A peptide synthesizer using a standard Fmoc protocol. Using 0.25mmol p-hydroxymethylphenoxymethyl polystyrene (HMP) resin as the starting resin, according to the amino acid sequence of the fusion peptide H9-V9, the peptide chain is extended from the carboxyl terminal to the amino terminal one by one. After the peptide chain is synthesized, , the resin containing the peptide chain was transferred to the cutting solution (composed of 0.25mL tartrate ethylenediamine, 9.5mL trifluoroacetic acid and 0.25mL deionized water), stirred at room temperature to cleave the peptide chain from the resin, and then used Filter through a G6 glass sand funnel, ...
Embodiment 2
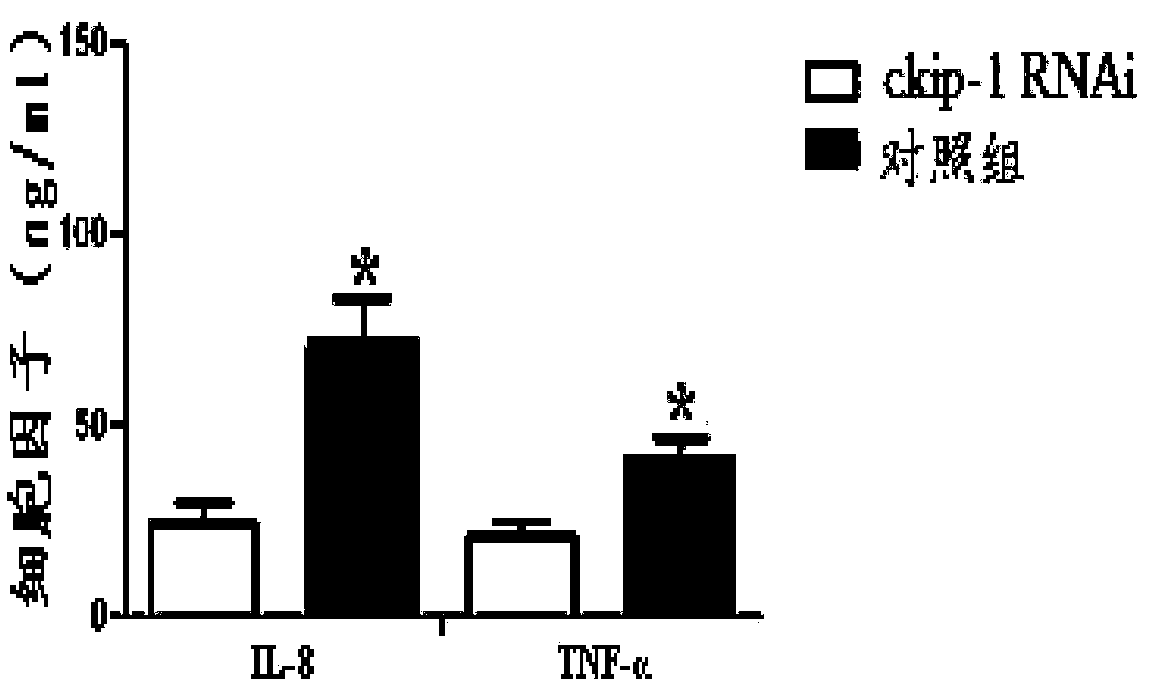
[0044] Example 2 Preparation of CKIP-1 RNAi complex
[0045] The nucleotide sequence of CKIP-1 RNAi is: 5'-CCTGAGTGACTATGAGAAG-3' (SEQ ID No.4). Under the condition of vortexing, to the aqueous solution containing CKIP-1 RNAi at a concentration of 200 μg / mL and citric acid-sodium citrate buffer at a concentration of 0.1 mol / L, add an equal volume of fusion peptide at a concentration of 50 μg / mL dropwise After the dropwise addition of the solution, continue to vortex for 60 minutes, and then stand still for 30 minutes to obtain the CKIP-1 RNAi complex targeting macrophages.
[0046] Drop the freshly prepared CKIP-1 RNAi complex on a 200-mesh copper grid, absorb it for 3 minutes, blot it with absorbent paper, dry it in the air for 30 seconds, negatively stain it with 1% uranyl acetate aqueous solution for 30 seconds, and use absorbent paper Blot dry, dry for 30 seconds, and observe with 80kV transmission electron microscope. The results are attached figure 1 As shown, the obt...
Embodiment 3
[0048] Example 3 Preparation of CKIP-1 RNAi complex
[0049] The nucleotide sequence of KIP-1 RNAi is: 5'-CCTGAGTGACTATGAGAAG-3' (SEQ ID No.4). Under the condition of vortexing, to the aqueous solution containing CKIP-1 RNAi at a concentration of 200 μg / mL and citric acid-sodium citrate buffer at a concentration of 0.1 mol / L, add an equal volume of fusion peptide at a concentration of 50 μg / mL dropwise After the dropwise addition of the solution, continue to vortex for 90 minutes, and then stand still for 60 minutes to obtain the CKIP-1 RNAi complex targeting macrophages.
[0050] Drop the freshly prepared CKIP-1 RNAi complex on a 200-mesh copper grid, absorb it for 3 minutes, blot it with absorbent paper, dry it in the air for 30 seconds, negatively stain it with 1% uranyl acetate aqueous solution for 30 seconds, and use absorbent paper Blot dry, dry for 30 seconds, and observe with 80kV transmission electron microscope. The results are attached figure 1 As shown, the obta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com