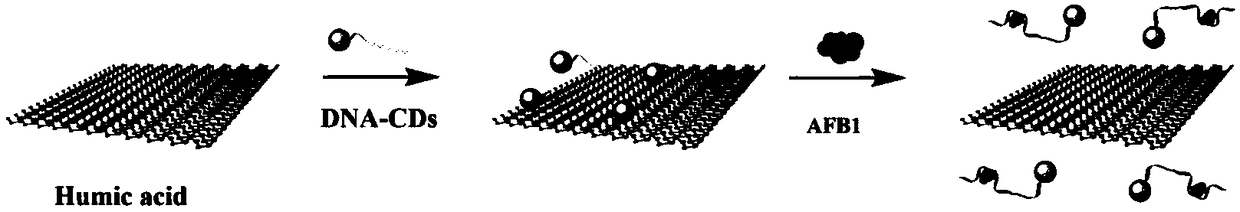
Rapid detection method for aflatoxin B1
A technology for the detection of aflatoxin and aflatoxin B1 is applied in the field of rapid detection of aflatoxin B1, which can solve the problems of cumbersome sample pretreatment procedures, expensive instruments, and low quantum yield, and is suitable for popularization and application. highly targeted effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1: Preparation of DNA-CDs complex
[0054] (1) Synthesis of amino-functionalized aptamer DNA:
[0055] The amino functionalized aptamer DNA was synthesized by conventional methods, and its sequence is: NH 2 - AAA AAA AAG TTG GGCACG TGT TGT CTC TCT GTG TCT CGT GCC CTT CGC TAG GCC CAC AC (SEQ ID NO. 1).
[0056] (2) Preparation of carbon dots (CDs):
[0057] Citric acid (1.0507 g) and ethylenediamine (335 μL) were dissolved in 10 mL of deionized water and stirred for 30 minutes, then transferred to a hydrothermal reactor (23 mL) and heated at 200 °C for 5 hours, then cooled naturally, A brown solution was obtained. The brown solution was purified by a dialysis membrane (1000MWCO) for 24 hours to remove small molecular substances in the solution, and freeze-dried by a freeze dryer to obtain dry carbon dot CDs.
[0058] (3) DNA-CDs complex preparation:
[0059] DNA-CDs complexes were prepared by cross-linking CDs and amino-functionalized DNA through EDC / NHS (ED...
Embodiment 2
[0060] Example 2: Drawing of AFB1 Detection Linear Equation
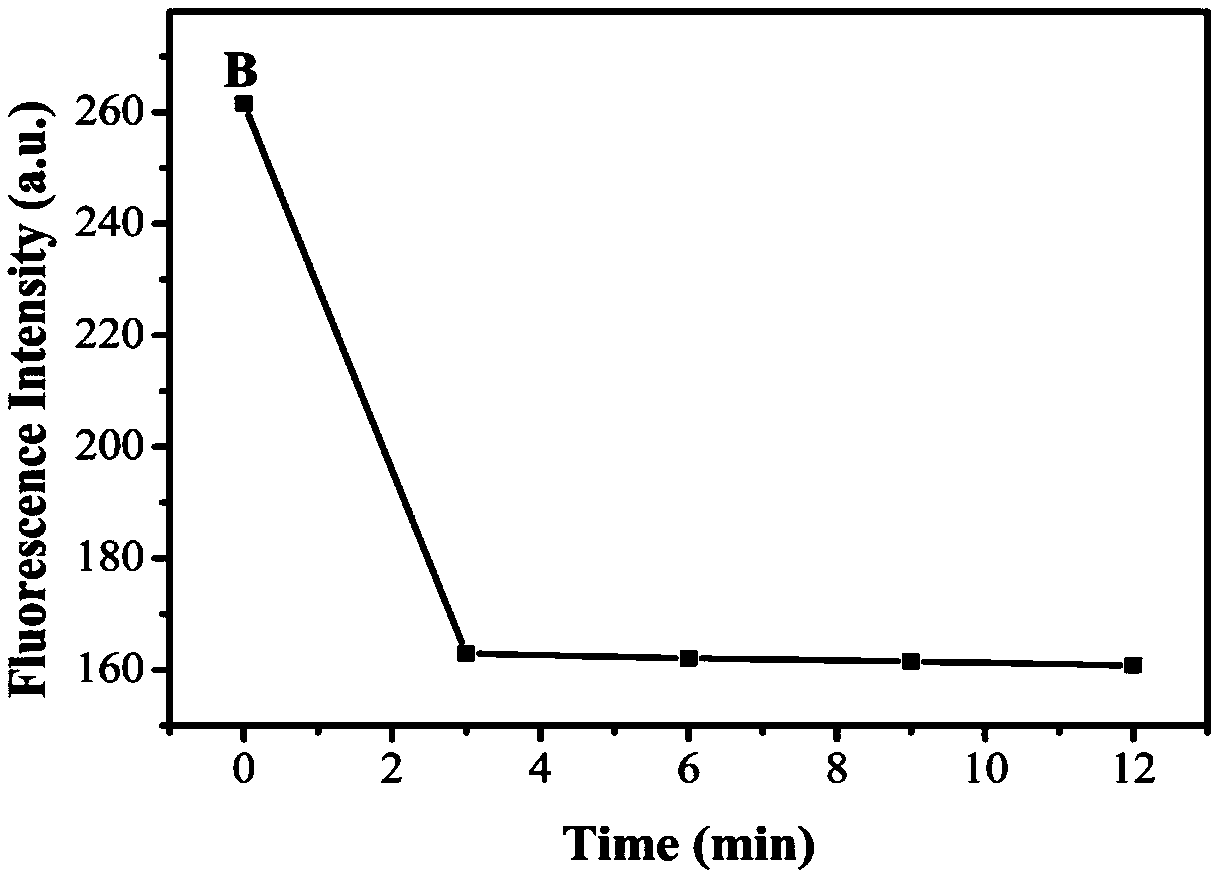
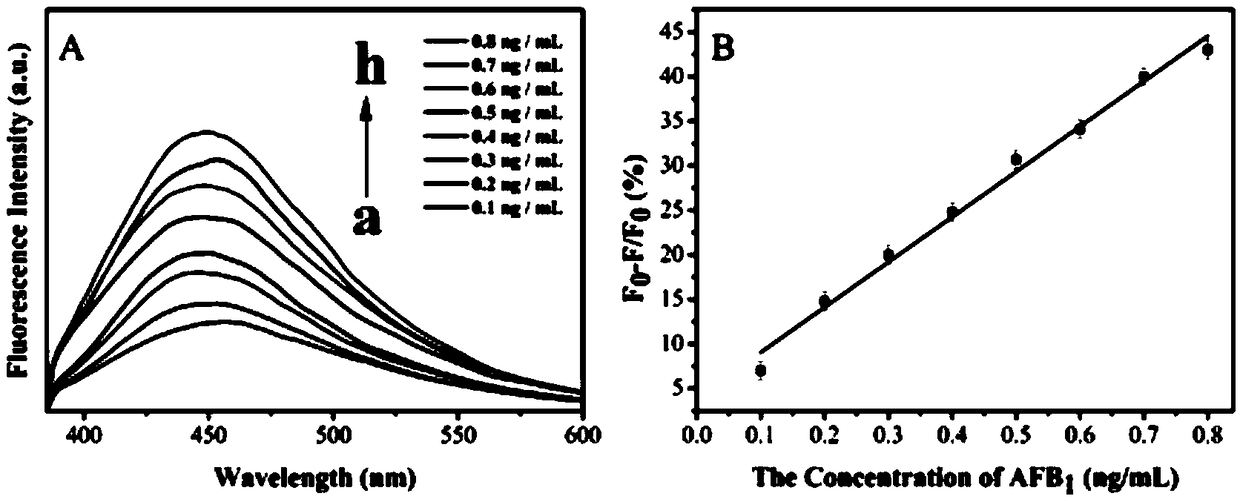
[0061] (1) Dissolve the DNA-CDs complex (prepared in Example 1) in sodium chloride solution (200 mM), make the concentration of the DNA-CDs complex 0.07 mM, adjust the pH to 7 with phosphate buffer, and incubate for 30 min, Under the excitation of 360nm light, measure its fluorescence intensity F 1 Add humic acid to make the concentration of humic acid 0.16mg / ml, react for 3min, measure its fluorescence intensity F under the excitation of 360nm light 0 ;
[0062] (2) After the reaction, add a series of concentration gradients (0.1ng / ml, 0.2ng / ml, 0.3ng / ml, 0.4ng / ml, 0.5ng / ml, 0.6ng / ml, 0.7ng / ml, 0.8ng / ml ml) AFB1 standard substance, under the excitation of 360nm light, measure its fluorescence intensity F 2 ; AFB1 fluorescence quenching rate I% is calculated according to the formula:
[0063] I%=(F 2 –F 0 ) / (F 1 -F 0 ), where F 0 Indicates the fluorescence intensity under 360nm excitation light when humic a...
Embodiment 3
[0066] Embodiment 3: the detection of aflatoxin B1 in the sample
[0067] (1) Dissolve the DNA-CDs complex (prepared in Example 1) in sodium chloride solution (200 mM), make the concentration of the DNA-CDs complex 0.07 mM, adjust the pH to 7 with phosphate buffer, and incubate for 30 minutes , Measure its fluorescence intensity F under the excitation of 360nm light 1 Add humic acid to make the concentration of humic acid 0.16mg / ml, react for 3min, measure its fluorescence intensity F under the excitation of 360nm light 0 ;
[0068] (2) Add the peanut oil sample to be tested after the reaction, and measure its fluorescence intensity F under the excitation of 360nm light 2 ; AFB1 fluorescence quenching rate I% is calculated according to the formula:
[0069] I%=(F 2 –F 0 ) / (F 1 -F 0 ), where F 0 Indicates the fluorescence intensity under 360nm excitation light when humic acid is added. f 1 Indicates the fluorescence intensity under 360nm excitation light without addin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com