A Quantitative Method of Fish Diversity Based on Edna and Its Sampling and Filtering Device
A filtration device and a variety of technologies, which can be used in sterilization methods, biological material sampling methods, support/immobilization methods for microorganisms, etc., which can solve the problems of high accuracy of analysis results and high experimental costs, affecting eDNA, cross-contamination and degradation, etc. problems, to achieve the effect of satisfying cloning and high-throughput sequencing, high specificity, and avoiding cross-contamination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
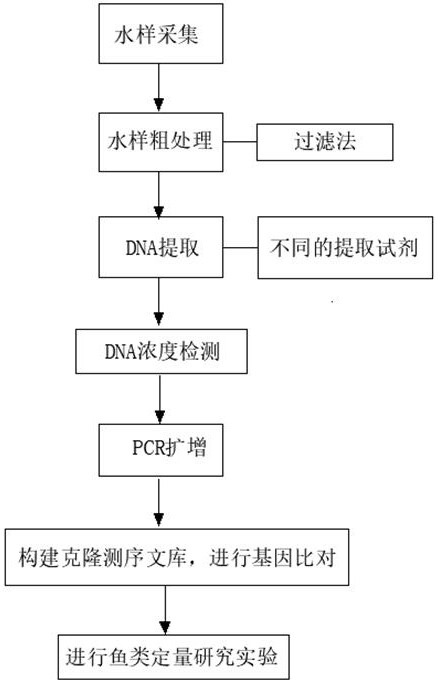
[0037] Embodiment 1: a kind of quantitative method and its sampling filtration device based on eDNA fish diversity, comprise the following steps:
[0038] S1. Using a sterilized sampling bottle, select the reservoir of the aquatic organism protection lake and its upstream and downstream for multiple sampling to collect 1L samples. Set a sampling point every one kilometer, and set 3 parallel samples at each sampling point. The sampling interval is 6 months;
[0039] S2. Use a sterilized eDNA-based quantitative sampling and filtration device for fish diversity to filter the collected samples, remove the microporous filter membrane after filtration, and place the microporous filter membrane in a refrigerator at -20°C cryopreservation in
[0040] S3, take out the microporous filter membrane and place it in a liquid nitrogen mortar and grind it, and pour the debris into a 2mL centrifuge tube for future use;
[0041] S4, take out the debris of the sample filter membrane, and use D...
Embodiment 2
[0053] Example 2: Based on Example 1, the difference is that the 7 sampling points in the upper and lower reaches of the Huairou Reservoir were intensively sampled, the sampling volume was 1L, and the collected samples used a sterilized eDNA-based fish Quantitative sampling and filtration device with various types of diversity, the filter membrane is placed in a liquid nitrogen mortar and crushed, and the DNA extraction experiment is carried out within 24 hours. The extracted DNA samples are put into an ultra-micro spectrophotometer for concentration detection, and the most covered species are selected Identify the universal primers with the highest accuracy, use the primers for diverse PCR amplification, take 2uL of the purified PCR product and connect it to the carrier, transform competent cells, and culture at a constant temperature at 37°C overnight. After cloning, pick 50 single colonies and use The primers were verified by colony PCR. The PCR product was detected by agaro...
Embodiment 3
[0055] Embodiment 3: Based on Examples 1 and 2, the difference is that 4 sampling points with the gene sequence of the broadfin carp are selected for sampling, and the collected samples are subjected to a DNA extraction experiment. According to the gene sequence of the BLAST comparison result, in Download the full gene sequence of Zaccoplatypus from the NCBI database, perform primer screening in the Primer5.5 software, obtain the upstream and downstream primer fragments of the target gene, and perform primer synthesis, using the collected samples as templates, using the target gene Primers are used for PCR amplification, and the recovered PCR products are subjected to TA cloning, and the target colonies are picked for plasmid extraction. The extracted plasmid is used as an absolute quantitative standard. The RealTimePCR reaction system is configured according to the DNA sample, and the RealTimePCR amplification experiment is carried out. Each sample is 3 repeated tests. Synthe...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com