Method for establishing tomato nuclear male sterile line with green stem marker
A male sterile line, tomato technology, applied in the field of creation of tomato nuclear male sterile line, can solve the problems of low breeding efficiency and low seed purity of tomato nuclear male sterile line
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
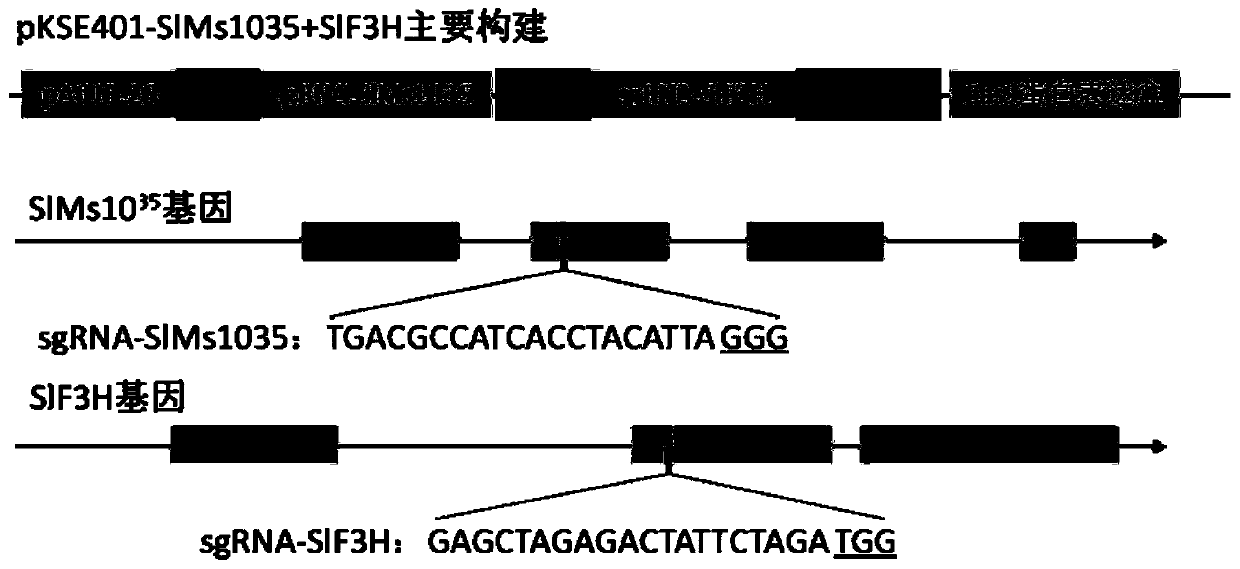
[0035] Embodiment 1 (SlMs10 35 , SlF3H and SlDFR target selection and construction of CRISPR / Cas9 gene editing vector)
[0036] 1. SlMs10 35 and SlF3H target selection
[0037] SlMs10 35 The gene numbers of the SlF3H gene in the tomato database are Solyc02g079810 and Solyc02g083860, both of which are located at the same end of the second chromosome of tomato. Using the online software CRISPR-PLANT (http: / / www.genome.arizona.edu / crispr / CRISPRsearch.html ) to design a target on each gene to construct a gene editing vector, the selected target sequence is as follows, and the underlined is the PAM region (prespacer sequence adjacent motif).
[0038] SlMs10 35 The target of the gene is located in the second exon region, and the target sequence is: TGACGCCATCACCTACATTA GGG (sequence 5);
[0039] The target of the SlF3H gene is located in the second exon region, and the target sequence is: GAGCTAGAGACTATTCTAGA TGG (sequence 6);
[0040] 2. Construction of recombinant vector...
Embodiment 2
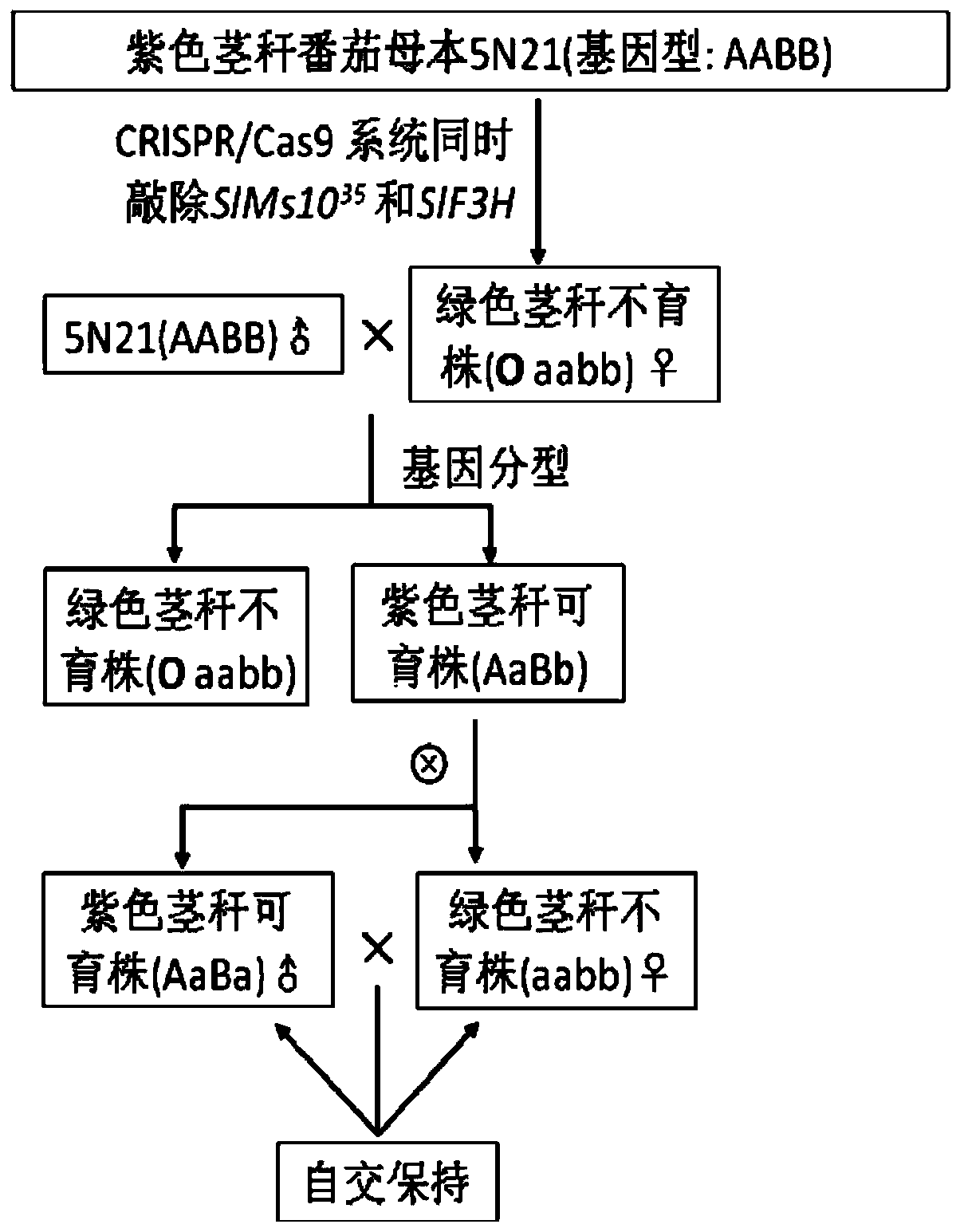
[0045] Embodiment 2 (transformation tomato)
[0046] The recombinant plasmid pKSE401-SlMs10 constructed in Example 1 35 +SlF3H was respectively introduced into tomato material 5N21 using Agrobacterium; the aseptic cotyledon of 5N21 was used as the transformation recipient, and Agrobacterium containing the recombinant plasmid was used to mediated transformation, and the complete regenerated plant was obtained through tissue culture after transformation;
[0047] Since the plants cannot produce anthocyanins after the SlF3H gene loses its function, the stems or young parts of the plants will not appear purple, so the plants with SlF3H gene editing can be selected;
[0048] Extract the genomic DNA of the plants selected above, and use specific primers to amplify the DNA containing SlMs10 35 and the gene sequence of the SIF3H target site; the amplified PCR product was TA-cloned to the T vector pEASY-T1 (Beijing Quanshijin Biotechnology Co., Ltd., CT101-01), and 5 single clones wer...
Embodiment 3
[0055] Embodiment 3 (slms10 35 Phenotype identification of slf3h-Cas9 plants)
[0056] slms10 obtained in Example 2 35 The slf3h-Cas9 plants were planted together with the wild-type 5N21 in the greenhouse, and the color of the stem, the shape of the flower and the development of pollen were observed; Figure 4 Planted seedlings are shown, wild-type 5N21 stems appear purple, while slms10 35 slf3h plant stems appear green; as Figure 5 As shown, when the plant grows to the second panicle of flowers, take the flowers that are fully open on the day of the plant for observation, and it can be seen that slms10 35 The flowers of the slf3h plant were smaller than the wild-type 5N21, and the stigma of the double mutant was obviously exposed due to the shortening of the stamens; the pollen was stained by Alexander's stain, and the pollen dyed red could be observed in the wild-type 5N21, while the slms10 35 Dyed pollen could not be observed in slf3h plants.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com