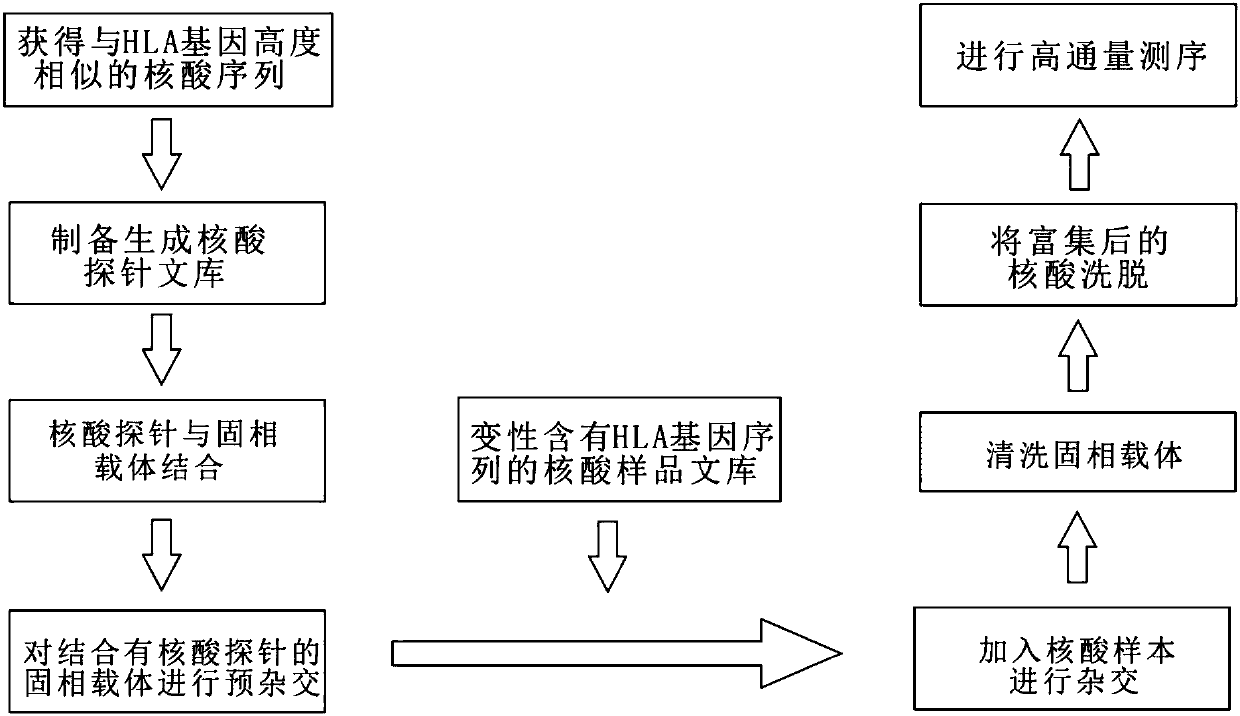
Method for targeted capture and sequencing of HLA gene sequences
A gene sequence and targeted capture technology, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms, can solve the problems of artificially synthesized probe sequences, low resolution and accuracy, and high cost, and achieve the realization of The effect of high abundance and high diversity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Example 1 Obtaining the double-stranded nucleic acid library after fragments and quality inspection
[0099] 1. Probe performance analysis
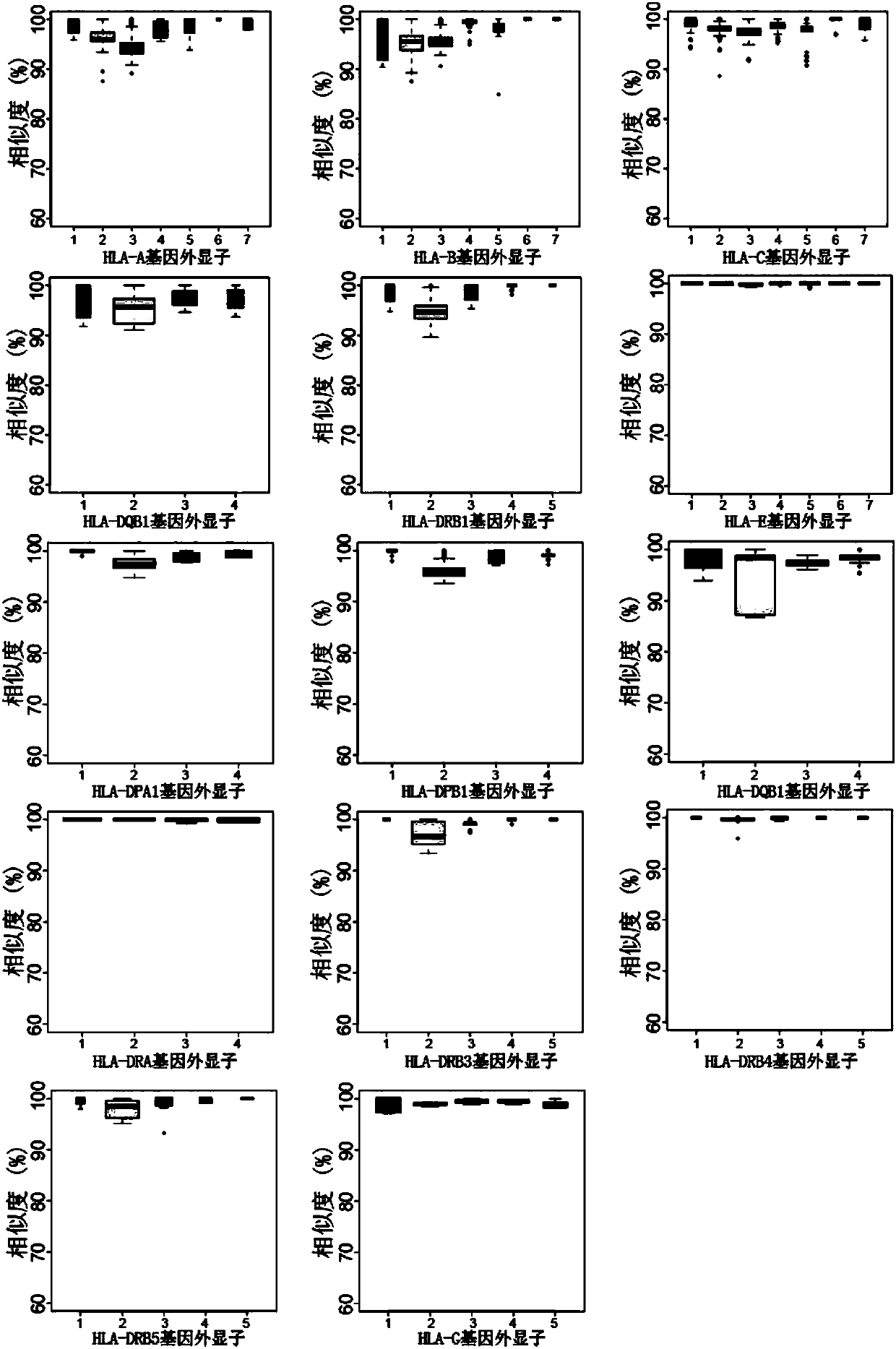
[0100] 1. Perform similarity analysis on the cDNA sequences of the selected 14 genes and the corresponding HLA gene sequences in the IMGT HLA database (version 3.26).
[0101] 2. Comparing the similarity in units of exons, the minimum similarity is 85%. This result shows that the cDNA sequences of these 14 genes in the database can all be captured. The results are shown in the appendix figure 2 .
[0102] 2. Preparation of double-stranded nucleic acid library:
[0103] 1. Mix 14 kinds of nucleic acid solutions according to the same number of molecules, including HLA-I family genes and HLA-II family genes, wherein the HLA-I family genes are HLA-A*01:01:01, B*08:01:01, C*16:01:01, E*01:01:01, G*01:01:02, HLA-II family genes are DPA1*02:01:01, DPB1*01 :01:01,DQA1*01:03:01,DQB1*03:02:01,DRA*01:01:01,DRB1*13:03:01,DRB3*01:01:02,DRB...
Embodiment 2
[0110] Example 2 Construction of DNA sample library
[0111] 1. The DNA samples, a total of 31 samples, were applied from the China Bone Marrow Bank. The DNA samples were samples with known HLA genotypes, and the method used was PCR-SBT.
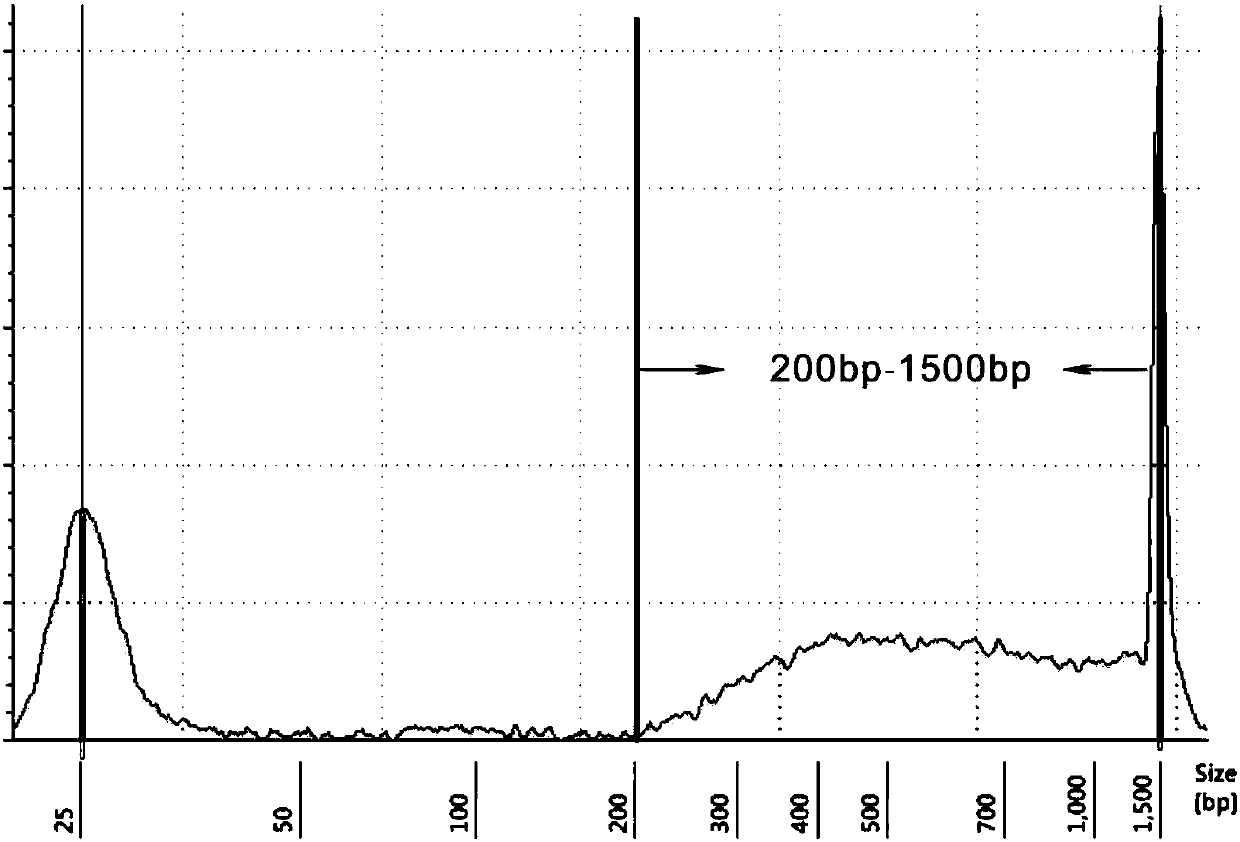
[0112] 2. Take 500ng of DNA sample and fragment the DNA with an ultrasonic instrument (Bioruptor). The conditions are 30s on / 90s off, 5 cycles, and the range of fragments generated is 300bp-600bp;
[0113] 3. Take 5 μl of the sonicated solution, add 1 μl of 6x loading Buffer, and check the size of the sonicated fragment by agarose gel electrophoresis, and the size is in the range of 300bp-600bp;
[0114] 4. For the remaining samples, use Ultra TM DNA Library Prep Kit for Illumina (E7645, NEB) was used to construct a sample DNA library, and the insert size range was 300-600bp.
Embodiment 3
[0115]Example 3 Enrichment of HLA gene sequences
[0116] 1. Preparation of single-stranded nucleic acid probe library:
[0117] 1. The preparation source of the single-stranded nucleic acid probe library is the double-stranded nucleic acid library in Example 1;
[0118] 2. Use alkaline denaturation method to denature the above double-stranded nucleic acid library and prepare at room temperature:
[0119] 24μl 500ng / μl DNA library
[0120] 12 μl 3M NaCl
[0121] 1.44 μl 10N NaOH
[0122] 3. Leave it at room temperature for 15 minutes to obtain a single-stranded nucleic acid probe library.
[0123] 2. Transfer the single-stranded nucleic acid probe to the solid-phase carrier membrane, so that the probe can be firmly combined with the positively charged nylon membrane:
[0124] 1. Cut about 0.3cm with scissors 2 Positively charged nylon membrane (NG0312, RPN3038, GE);
[0125] 2. Use a pipette gun to transfer the denatured solution containing the single-stranded nucleic a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com