Septin 9 gene methylation detection method and kit
A detection kit and methylation technology, which is applied in the field of human gene methylation detection, can solve the problems of reducing the sensitivity of PCR and subsequent analysis techniques, single Septin9 gene fragments, severe bisulfite conversion conditions, etc., and achieve detection results Accurate and reliable, prevent false positive results and false negative results, and improve the effect of detection specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
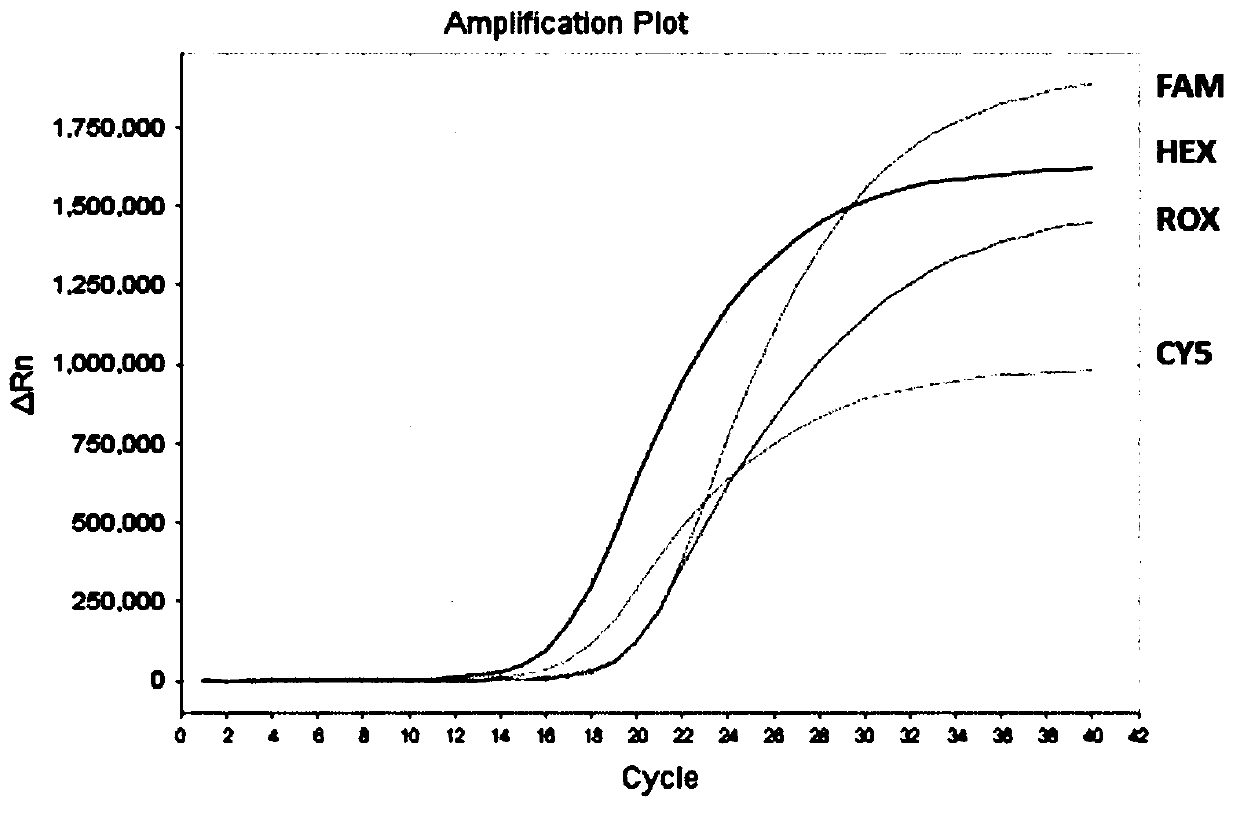
[0054] Example 1: A method and kit for Septin 9 gene methylation detection
[0055] A method and kit for detecting the methylation of Septin 9 gene, the detection method involved is to use TET enzyme to oxidize 5caC in 5mC and 5hmC in the DNA to be tested, then reduce 5caC to dihydro DHU with pyridine borane, and then use Specific primers and probes designed for gene methylation establish a multiplex fluorescent PCR system for PCR amplification, during which DHU is converted into T, and the methylation status of the Septin 9 gene is judged according to the fluorescent signal of the multiplex fluorescent PCR reaction; The kit includes TET oxidation buffer, TET enzyme, fluorescent PCR pre-reaction solution, polymerase, negative quality control, and positive quality control. The preparation of the Septin 9 gene methylation detection kit includes the following steps:
[0056] (1) Prepare TET oxidation buffer solution according to the formula of TET oxidation buffer solution of th...
Embodiment 2
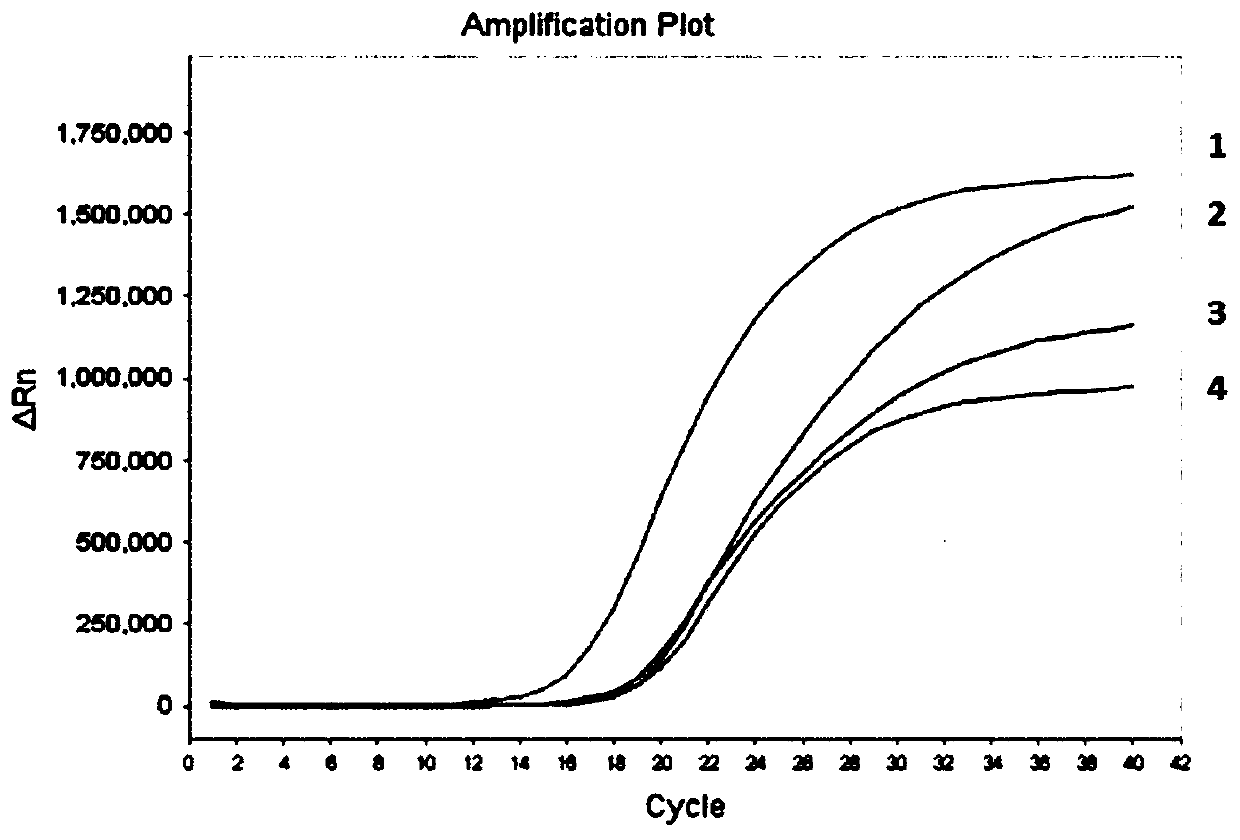
[0093] Embodiment 2: sensitivity analysis experiment
[0094] (1) Selection of samples to be tested
[0095] In this embodiment, methylated human genomic DNA is selected as the sample to be tested. In order to simulate the length of free DNA fragments, the methylated human genomic DNA as the sample to be tested was ultrasonically treated in this experiment to fragment it.
[0096] (2) Transformation of DNA samples
[0097] Using a method and kit for detecting methylation of the Septin 9 gene obtained in Example 1, perform TET-assisted pyridine borane treatment on the fragmented methylated human genomic DNA according to the detection steps in Example 1 to obtain converted DNA .
[0098] (3) Multiplex fluorescent PCR detection
[0099] The converted DNA concentration obtained according to the method described in Example 1 was adjusted to 10ng / ml, 1ng / ml, 100pg / ml, 50pg / ml, 25pg / ml, 10pg / ml, and 1pg / ml, and used as DNA templates in Examples The method and kit for detecting t...
Embodiment 3
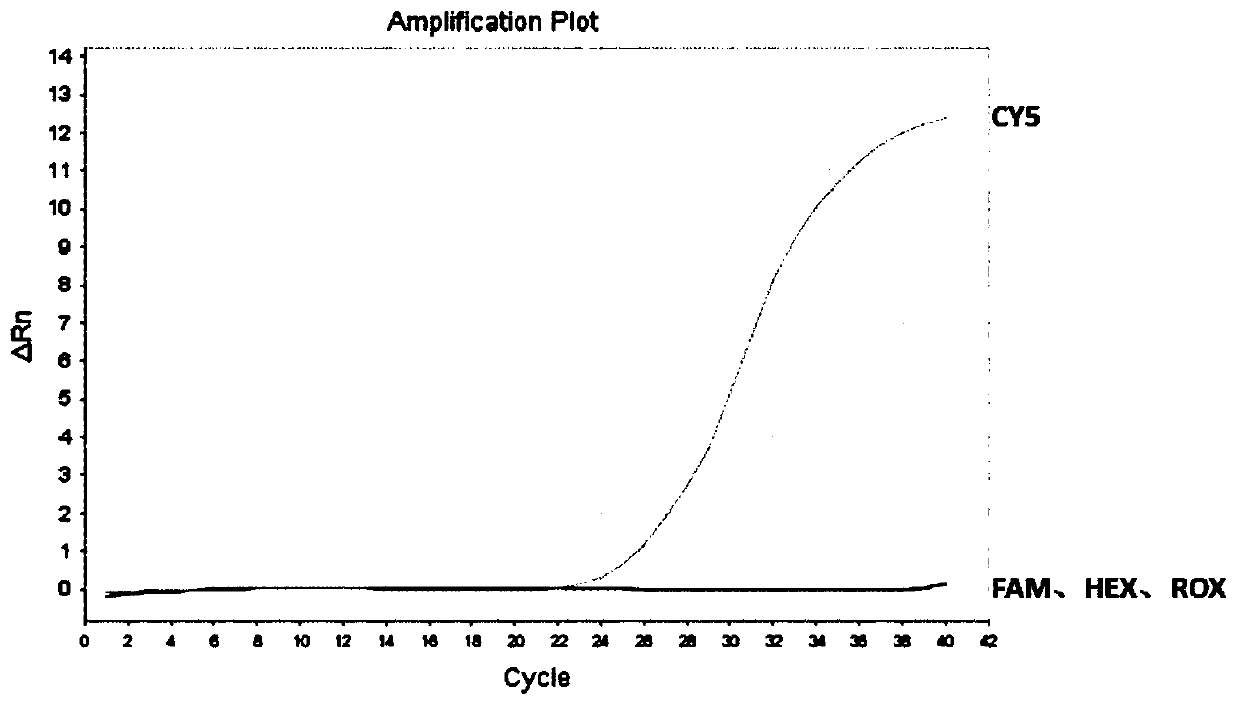
[0103] Embodiment 3: specificity analysis experiment
[0104] (1) Selection of samples to be tested
[0105] In this embodiment, unmethylated human genomic DNA, methylated human genomic DNA, 8 DNA samples from healthy individuals, and 8 DNA samples that were positive for Septin 9 methylation determined by bisulfite sequencing were selected as Sample to be tested. Wherein, non-methylated human genome DNA and methylated human genome DNA are fragmented by ultrasonic treatment.
[0106] (2) Transformation of DNA samples
[0107] Using the method and kit for detecting the methylation of the Septin 9 gene described in Example 1, according to the detection steps in Example 1, the above-mentioned samples to be tested were treated with TET-assisted pyridine borane to obtain converted DNA.
[0108] (3) Multiplex fluorescent PCR detection
[0109] The transformed DNA obtained above is used as a DNA template to perform multiple fluorescent PCR detection according to the detection step...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com