Alkaline protease PA3 and its coding gene and application thereof
A protease and alkaline technology, applied in the field of agricultural biology, can solve problems such as differences in amino acid results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
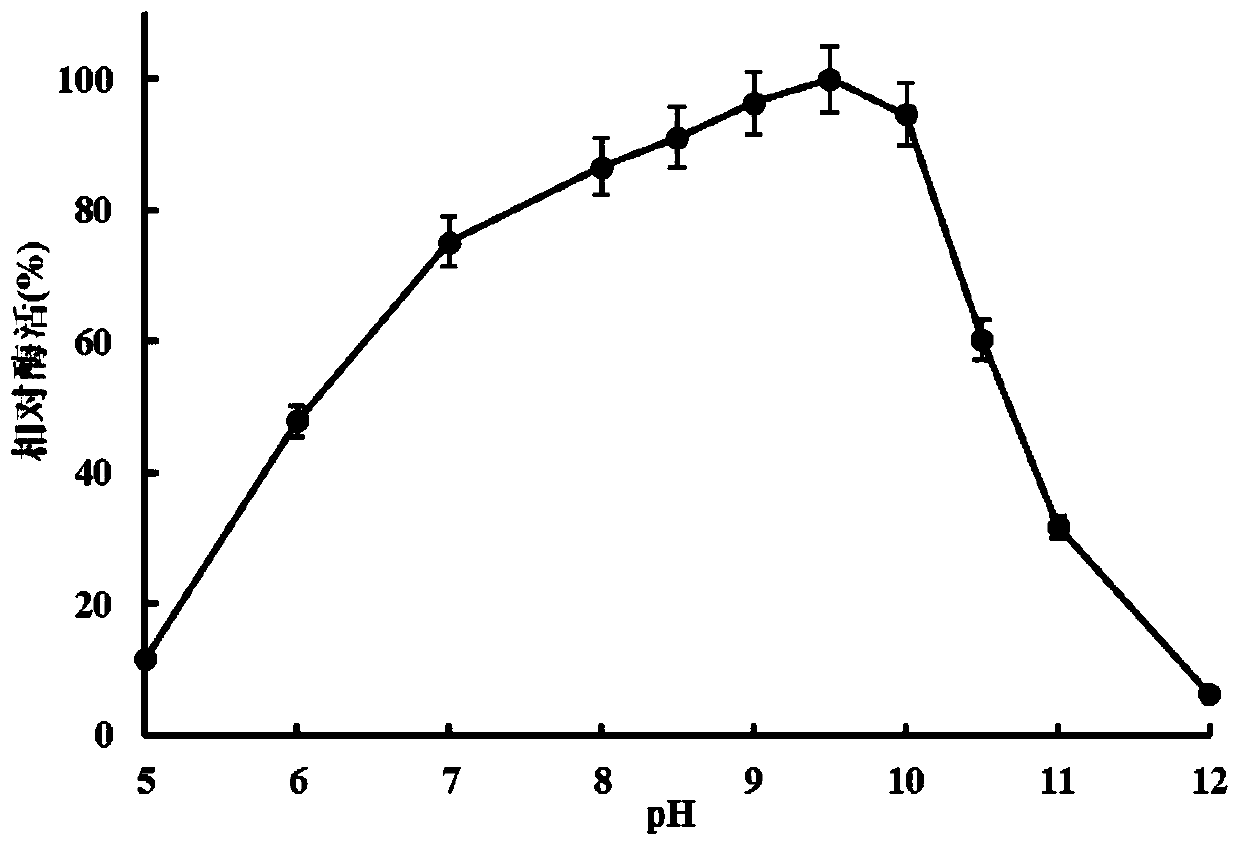
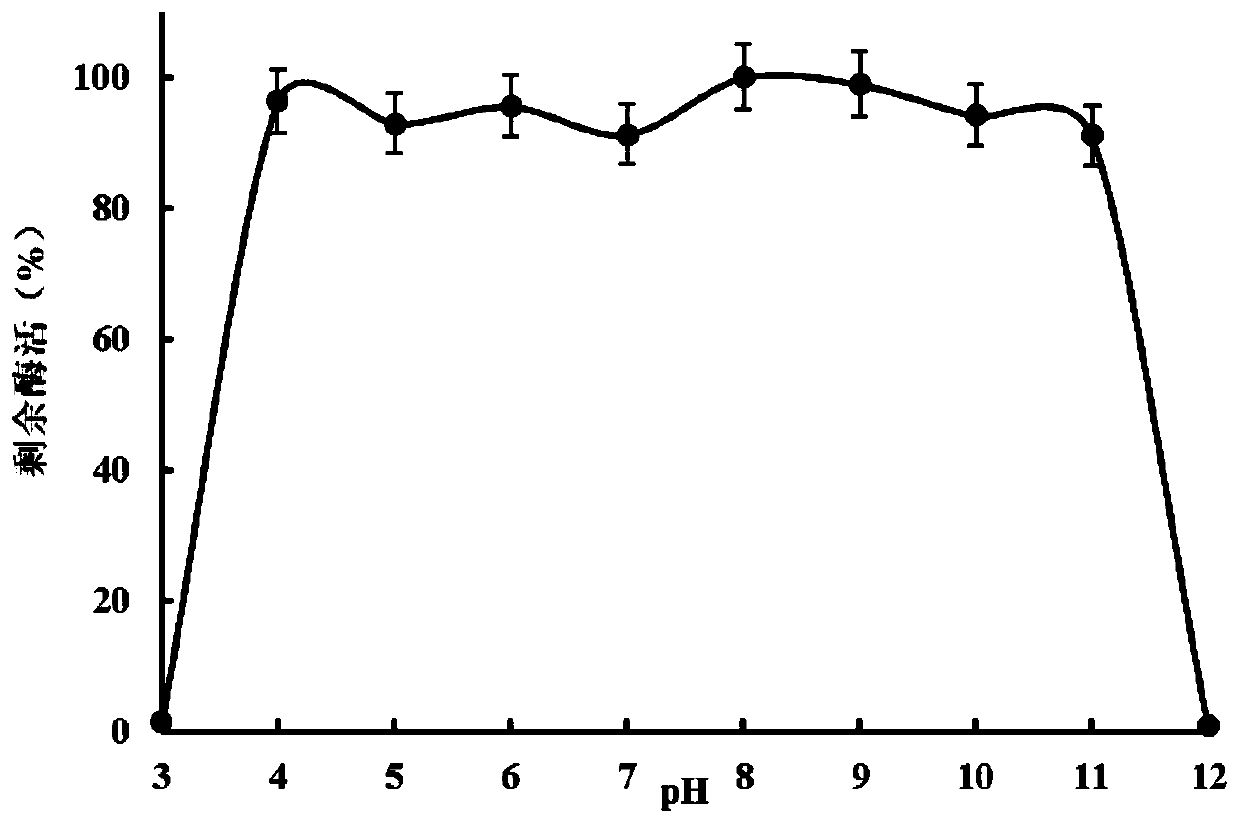
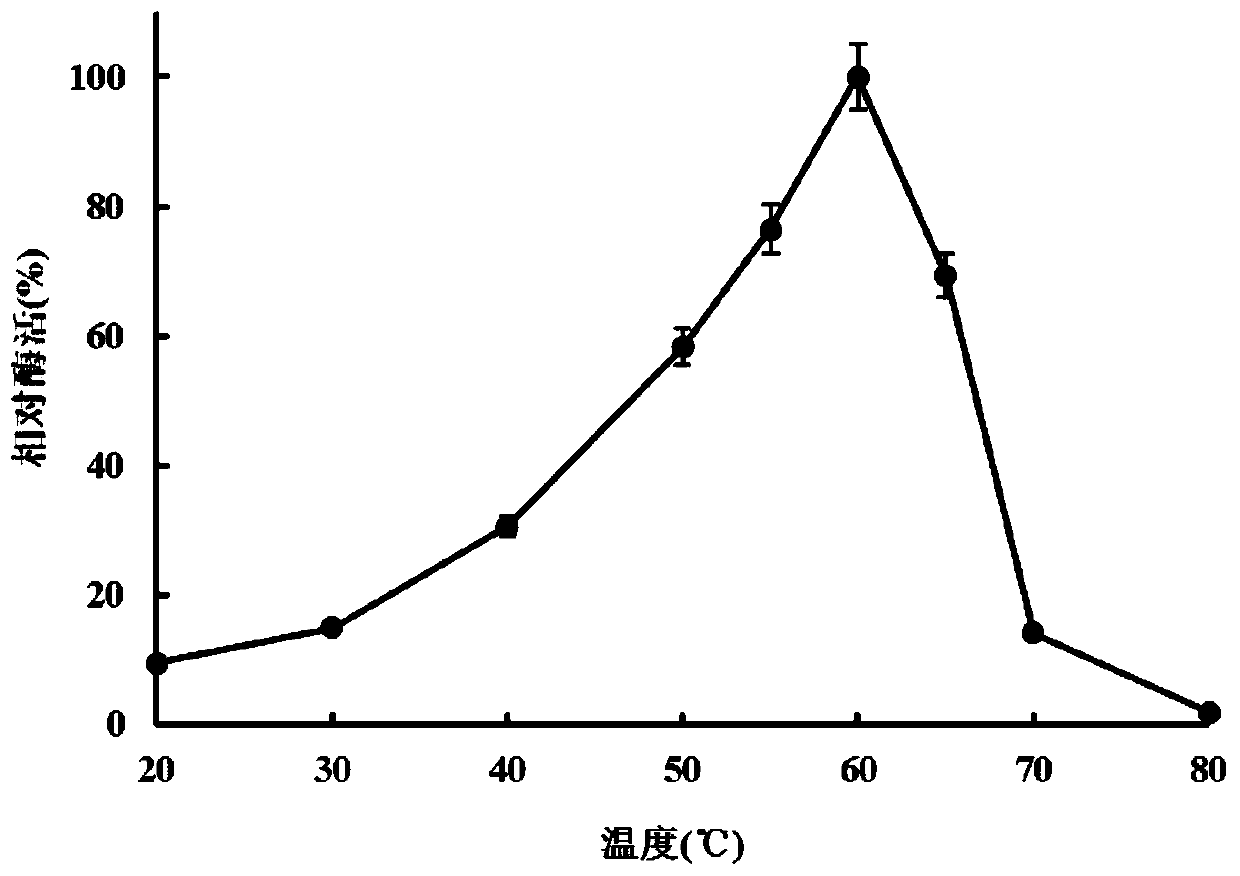
Image
Examples
Embodiment 1
[0052] Embodiment 1 obtains alkaline protease gene pa3 and constructs expression vector
[0053] According to the protein sequence published in Genbank (serial number: CEJ93023.1), codon optimization was performed on it, and the optimized gene was obtained by chemical synthesis, numbered pa3.
[0054] The gene pa3 is connected to the pUC57 carrier, and the present invention realizes the connection of the target gene pa3 and the expression vector pPIC9r through the method of homologous recombination, and completes the construction of the expression vector.
[0055] First, through the PCR method, specific primers are used to introduce homologous segments into the target gene.
[0056] Table 1 pa3-specific primers
[0057]
[0058] Note: The underline marks the overlapping sequence homologous to the vector pPIC9r
[0059] Using the recombinant plasmid pUC57-pa3 as a template, carry out PCR amplification reaction through specific primers; at the same time, use restriction end...
Embodiment 2
[0062] The construction of embodiment 2 alkaline protease PA3 engineering strains
[0063] A large number of recombinant pPIC9-pa3 plasmids were extracted, and linearized using restriction endonuclease BglII to linearize the system.
[0064] After digestion at 37°C for 2 hours, 10 μL of the digestion product was subjected to agarose gel electrophoresis to confirm whether the digestion was complete, and then the digestion product was recovered and purified using the Omega Gel Extraction kit. The purified and recovered linearized product was electrotransformed into a competent Pichia pastoris (electrotransformer parameters: Fungus, PIC). The bacteria solution after electrotransfer was spread on the MD plate and incubated at 30°C for 48h. After transformants grew on the MD plate, 36 single clones were picked from the MD plate with a sterilized toothpick, placed on the milk double-layer screening plate, and cultured at 30° C. for 72 hours. According to whether there are hydrolys...
Embodiment 3
[0065] Embodiment 3 prepares recombinant alkaline protease PA3
[0066] (1) Shake flask horizontal expression of recombinant alkaline protease PA3
[0067] The selected transformants with higher enzyme activity were inoculated in 30 mL of YPD liquid medium, and cultured at 30° C. and 200 rpm in a shaker for 48 hours to obtain seed liquid. Then the seed solution was transferred to 300mL BMGY medium according to the inoculation amount of 1%, and cultivated in a shaker at 30°C and 200rpm for 48h, and then the bacteria were transferred to 200mL BMMY medium, and cultured in a shaker at 30°C and 200rpm After culturing for 48 hours, 0.5% (V / V) methanol was added every 24 hours during this period.
[0068] (2) Purification of recombinant alkaline protease PA3
[0069] After the fermentation process, the bacterial liquid was centrifuged at 12000rpm for 10 minutes to remove the bacterial cells and other insoluble matter, and the supernatant was collected, which was the crude enzyme li...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com