Engineering bacteria producing l-rhamnose and its construction method and application
A technology of rhamnose isomerase and rhamnose, which is applied in the field of genetic engineering, can solve the problems of many by-products, high cost, and affecting the yield and output of rhamnose, and achieve the effect of avoiding hydrolysis steps and reducing the burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
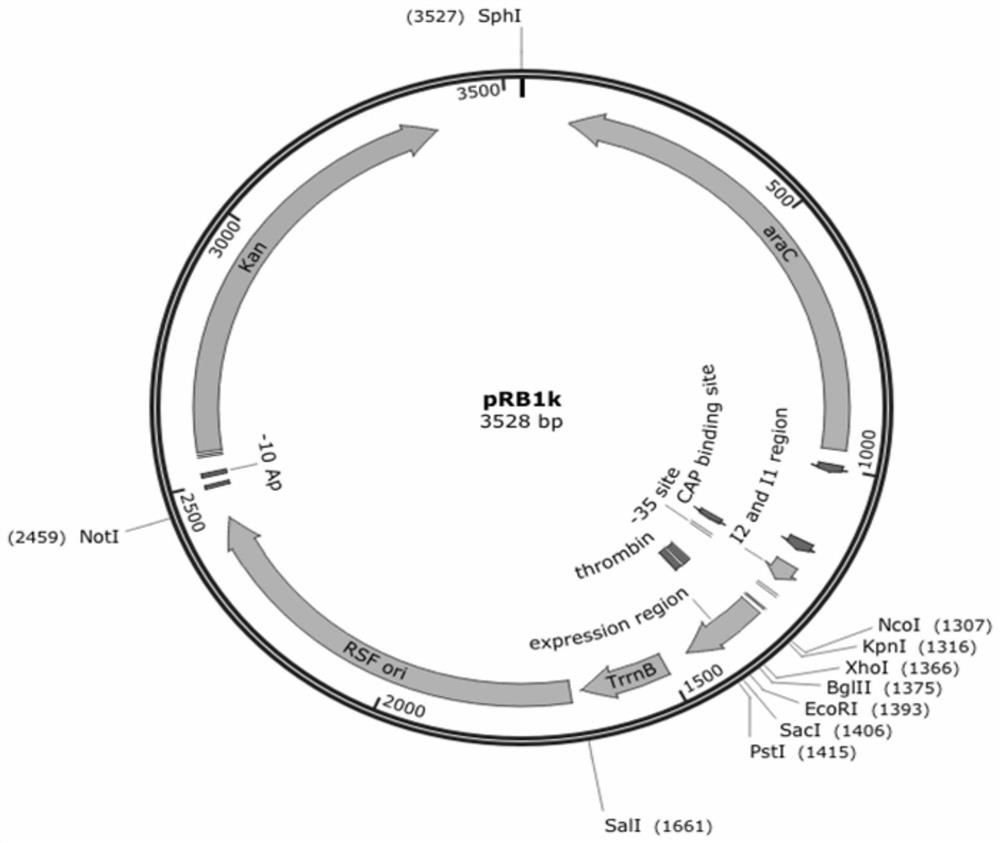
[0098] Example 1. Construction of recombinant plasmids co-expressing rhamnose-1-phosphate aldolase, fructose-1-phosphatase and L-rhamnose isomerase
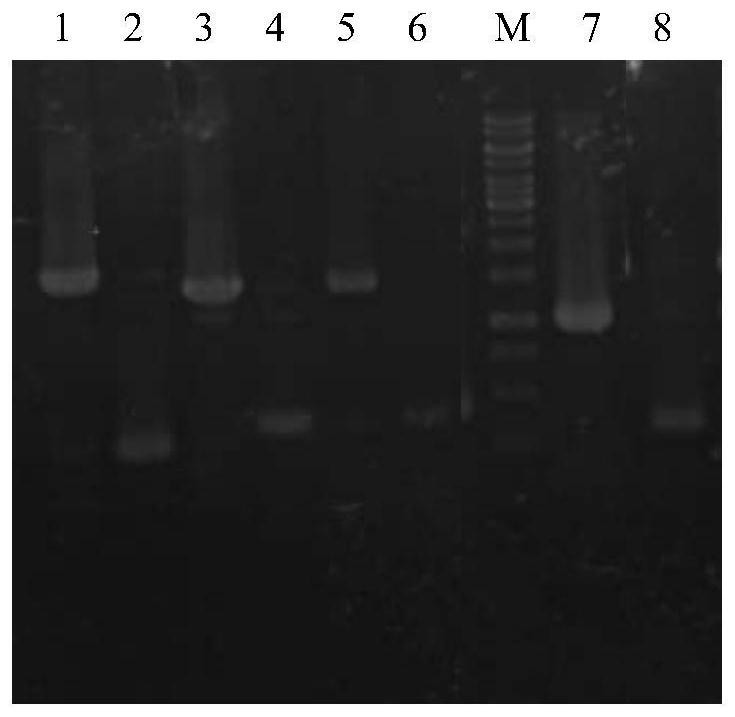
[0099] 1. Using the Escherichia coli K12 genome as a template, the primer pair P1 and P2 are used for PCR amplification, and the gene encoding fructose-1-phosphatase (yqaB) is obtained by PCR amplification. The PCR conditions were as follows: 98°C, 2min; 98°C, 20sec, 55°C, 20sec, 72°C, 30sec (30 cycles); 72°C, 5min. Detected by 1% agarose gel electrophoresis, the fragment size is about 570bp, which is consistent with the target fragment size, and the yqaB gene fragment is recovered.
[0100] 2. Using the Escherichia coli K12 genome as a template, the primer pair P3 and P4 were used for PCR amplification, and the coding gene (rhaA) of L-rhamnose isomerase was obtained by PCR amplification. The PCR conditions were as follows: 98°C, 2min; 98°C, 20sec, 55°C, 20sec, 72°C, 45sec (30 cycles); 72°C, 5min. Detected by 1% agarose gel ele...
Embodiment 2
[0114] Embodiment 2, construct the recombinant plasmid that co-expresses methylglyoxal synthase and glycerol dehydrogenase
[0115] 1. Construction of recombinant plasmid pFU54
[0116] (1) Using the Escherichia coli K12 genome as a template, the primer pair P9 and P10 were used for PCR amplification, and the coding gene (gldA) of glycerol dehydrogenase was obtained by PCR amplification. The PCR conditions were as follows: 98°C, 2min; 98°C, 20sec, 55°C, 20sec, 72°C, 45sec (30 cycles); 72°C, 5min. Detected by 1% agarose gel electrophoresis, the fragment size was about 1100bp, which was consistent with the target fragment size, and the gldA gene fragment was recovered.
[0117] (2) After the pBAD vector was double-digested with XhoI and EcoRI, a large vector fragment with a size of about 4000 bp was recovered.
[0118] (4) The gldA gene fragment recovered in step (1) and the large vector fragment were used Gibson method (Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, ...
Embodiment 3
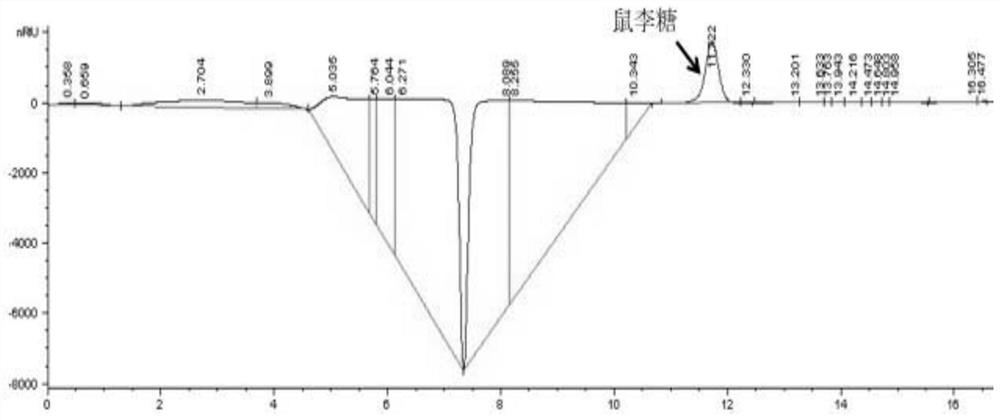
[0133] Embodiment 3, construct the host bacterium AODT that produces L-rhamnose
[0134] Chromosome editing of wild-type Escherichia coli K12 to knock out the aldehyde dehydrogenase A gene (aldA), L-1,2-propanediol oxidoreductase gene (fucO), and NADPH-dependent aldehyde reductase gene of wild-type Escherichia coli K12 (yqhD) and triose phosphate isomerase gene (tpiA), to obtain Escherichia coli mutant AODT. In this example, the P1 phage-mediated transfection method was used to construct the Escherichia coli mutant AODT, and the specific steps were as follows:
[0135] (1) Obtain the P1 of the donor bacteria: the donor bacteria BW25113△aldA::Kan (National Institute of Genetics (NIG, Japan), NIG number is JW1412), BW25113△fucO::Kan (National Institute of Genetics (NIG, Japan), NIG number JW2770), BW25113△yqhD::Kan (National Institute of Genetics (NIG, Japan), NIG number JW2978) and BW25113△tpiA::Kan (National Institute of Genetics (NIG , Japan), NIG No. JW3890) were inoculate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com