Extraction reagent and extraction method for fermented bean curd microorganism flora DNA
A technology of microorganisms and fermented bean curd, which is applied to the extraction reagent and extraction of fermented bean curd microbial community DNA: 1) In the field of microbial separation and recovery in fermented bean curd, it can solve the problems of unsuitable fermented bean curd microbial community
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] A set of reagents for extracting microbial DNA of fermented bean curd, including cell separation liquid, cell washing liquid, cell lysing liquid, PCR inhibitory substance precipitation liquid and cleaning liquid.
[0045] Preparation of cell separation solution: weigh 1.36g KH 2 PO 4 , 3.58 g Na 2 HPO 4 12H 2 O, 5.84g NaCl, 0.07g KCl, and 800ml of deionized water were added. After stirring evenly with a glass rod, the pH value was adjusted to 7.5, and the volume was adjusted to 1L by adding deionized water.
[0046] Preparation of cell washing solution: weigh 1.36g KH 2 PO 4 , 3.58 g Na 2 HPO 4 12H 2 O, add 1ml Tween 20 and add 800ml deionized water, stir evenly with a glass rod, adjust the pH value to 7.5, add deionized water to make up to 1L.
[0047] Preparation of cell lysate: Weigh 24.2g Tris (trishydroxymethylaminomethane), 74.5g KCl, 0.3g EDTA, add 800ml deionized water, stir evenly with a glass rod, add 5ml SDS, 5ml Triton x- 100, adjust the pH value t...
Embodiment 2
[0051] Extract commercialized fermented bean curd microbial DNA from different manufacturers, and use the extraction reagent provided in Example 1 to extract, a total of 12 fermented bean curd samples, the steps are as follows:
[0052] (1) 10g fresh fermented bean curd sample is mashed sample with Mixer Mill MM 400 stirring homogenizer of RETSCH company;
[0053] (2) Weigh 10 g of the crushed fermented bean curd sample, add 1 g of tungsten carbide beads with a diameter of 16 mm, and add 50 ml of cell separation medium SFS buffer, shake at room temperature for 15 minutes on a shaker at 120 rpm;
[0054] (3) Let the appeal sample stand at room temperature for 5 minutes, centrifuge at 13000×g for 1 minute, collect the precipitate, pour off all the supernatant, and weigh about 5 g of the precipitate to proceed to the next step;
[0055] (4) Add 25mL of cell washing solution SFW buffer, shake on a shaker at 120 rpm at room temperature for 10 minutes, let stand at room temperature ...
Embodiment 3
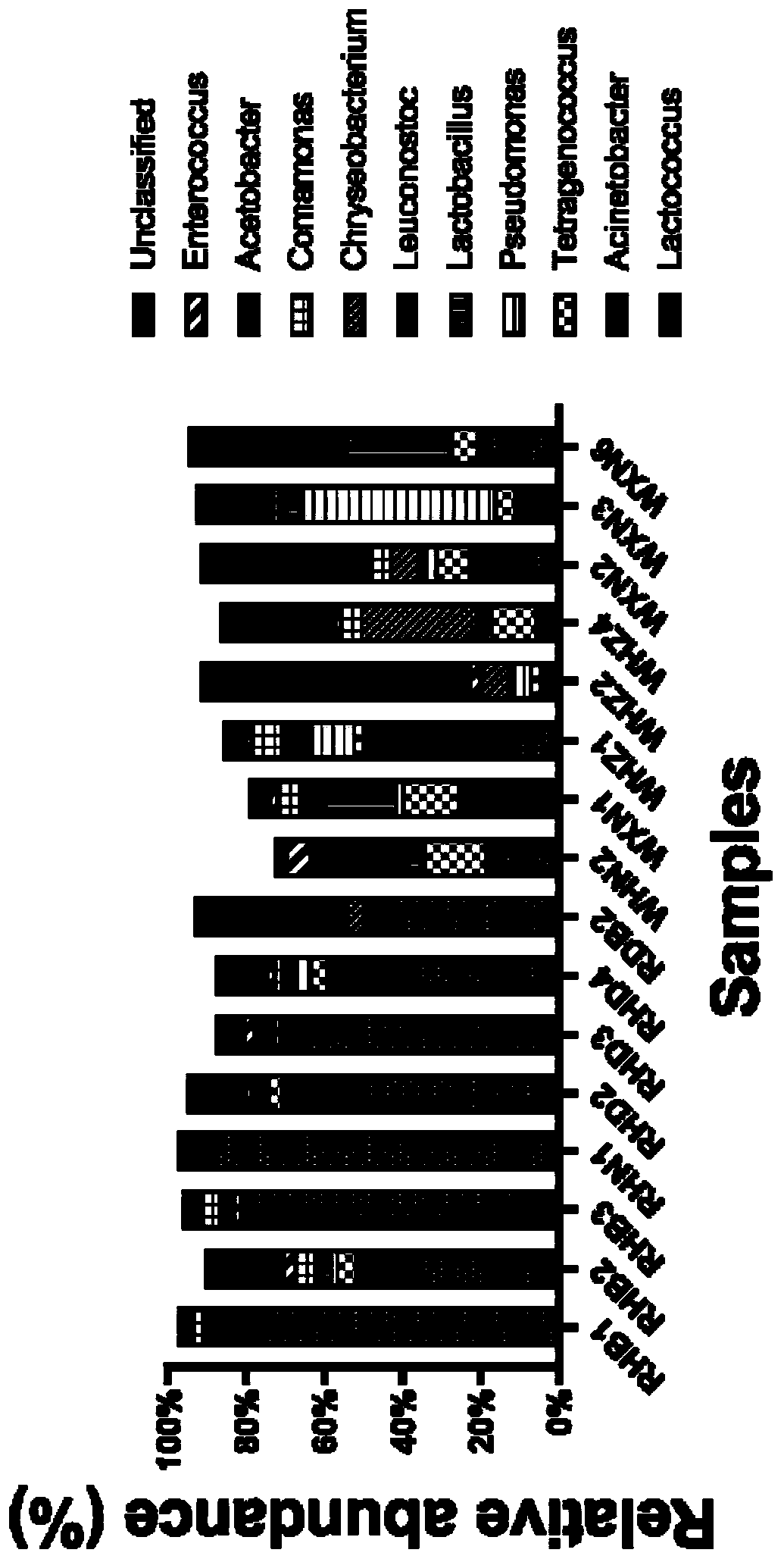
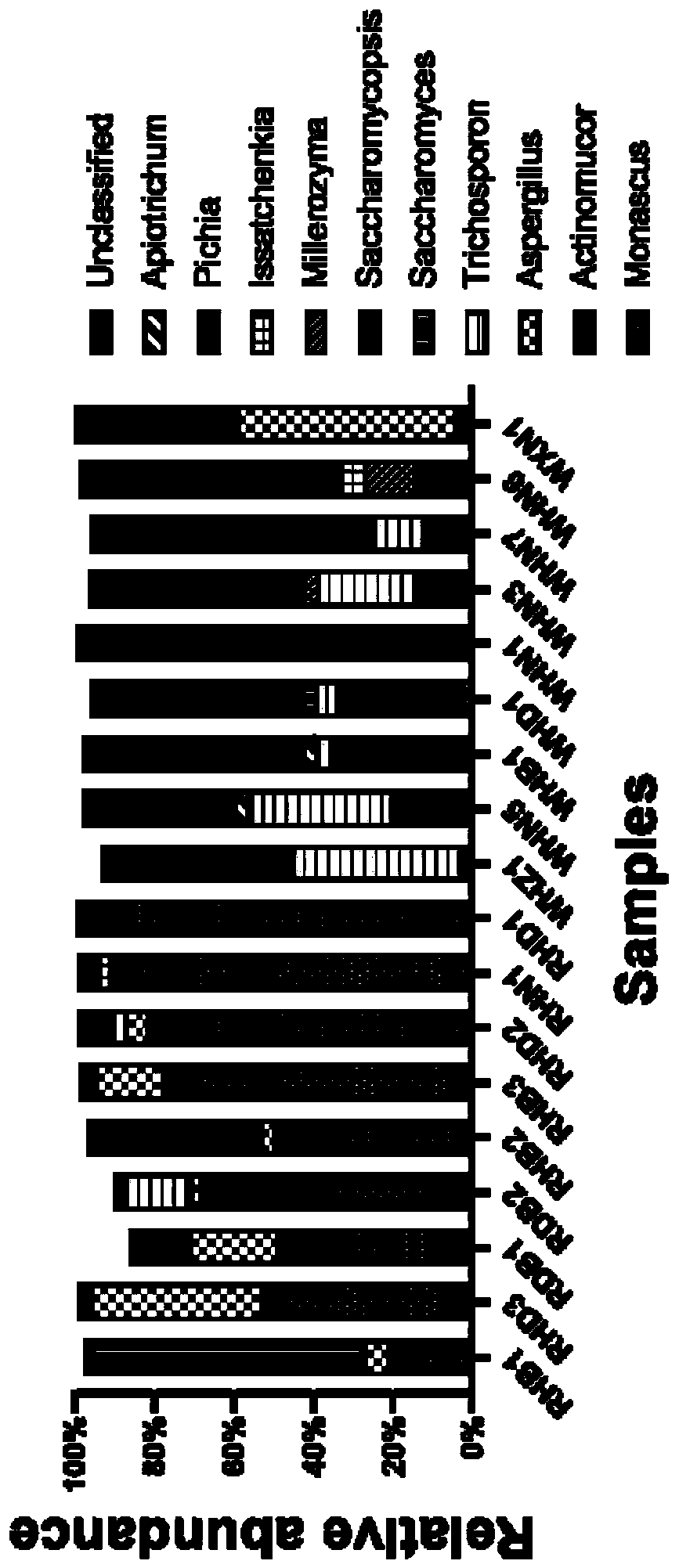
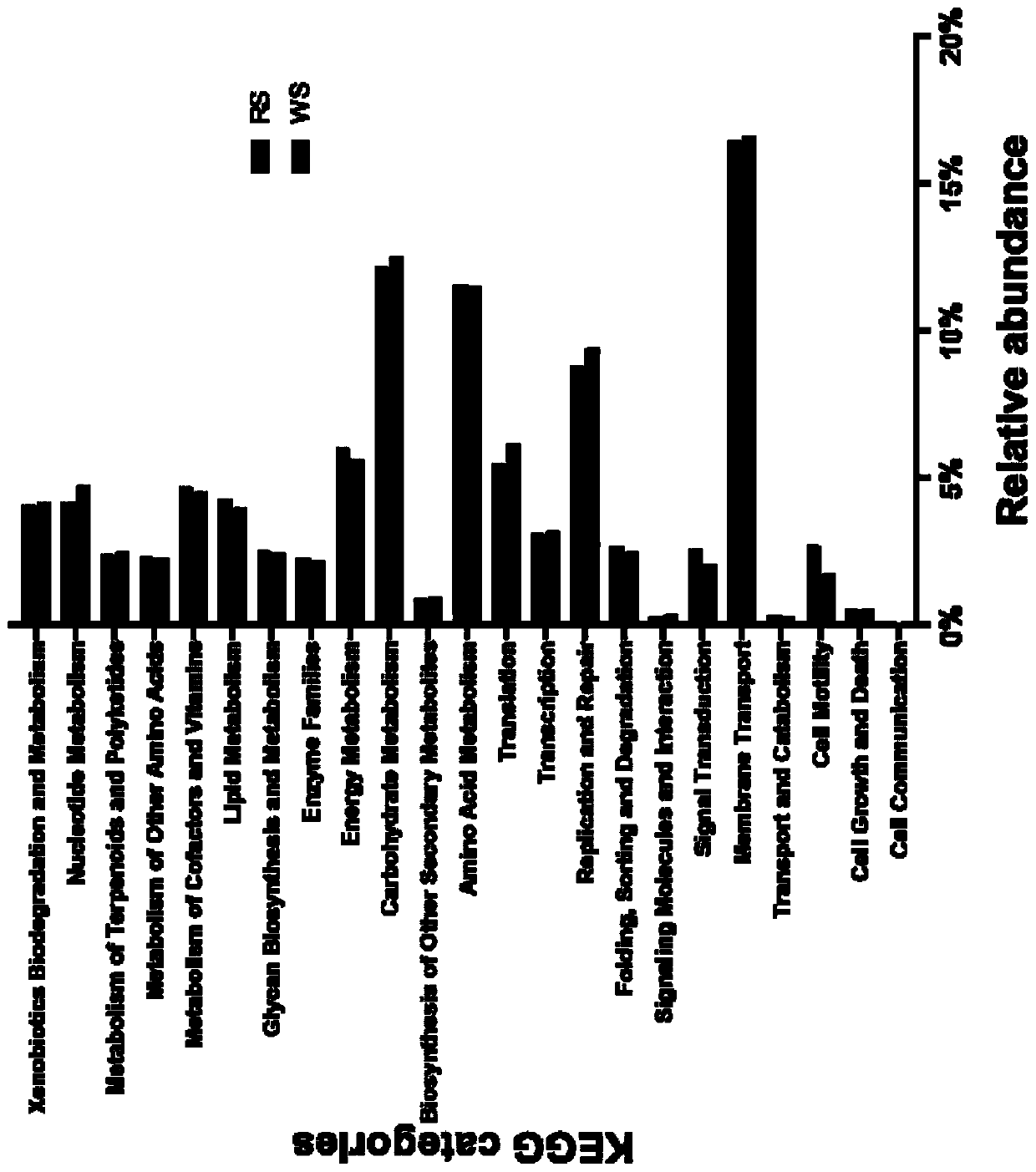
[0070] Six commercial red bean curd and six white bean curd were collected, their DNA was extracted, 16S rRNA gene was amplified by PCR, and the bacterial community composition of the bean curd samples was identified by high-throughput sequencing. Adopt the extraction reagent that embodiment 1 provides to extract, altogether 12 fermented bean curd samples, the steps are as follows:
[0071] (1) The DNA extraction method is the same as in Example 2, followed by PCR amplification of the 16S rRNA gene and high-throughput sequencing of the amplicon and bioinformatics analysis method;
[0072] (2) Take 1 μl of DNA as a template to amplify the V4 region of the 16S rRNA gene. 515F(5'-GTG CCA GCM GCC GCGGTA A-3')+806R(5'-GGA CTA CHV GGG TWT CTA AT-3') was used as primers for PCR amplification. The total volume of the PCR amplification system is 50 μl, and the composition of the reaction system is: 1 μl of upstream and downstream primers (concentration 10 μM), 2 μl of 10×Extag reactio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com