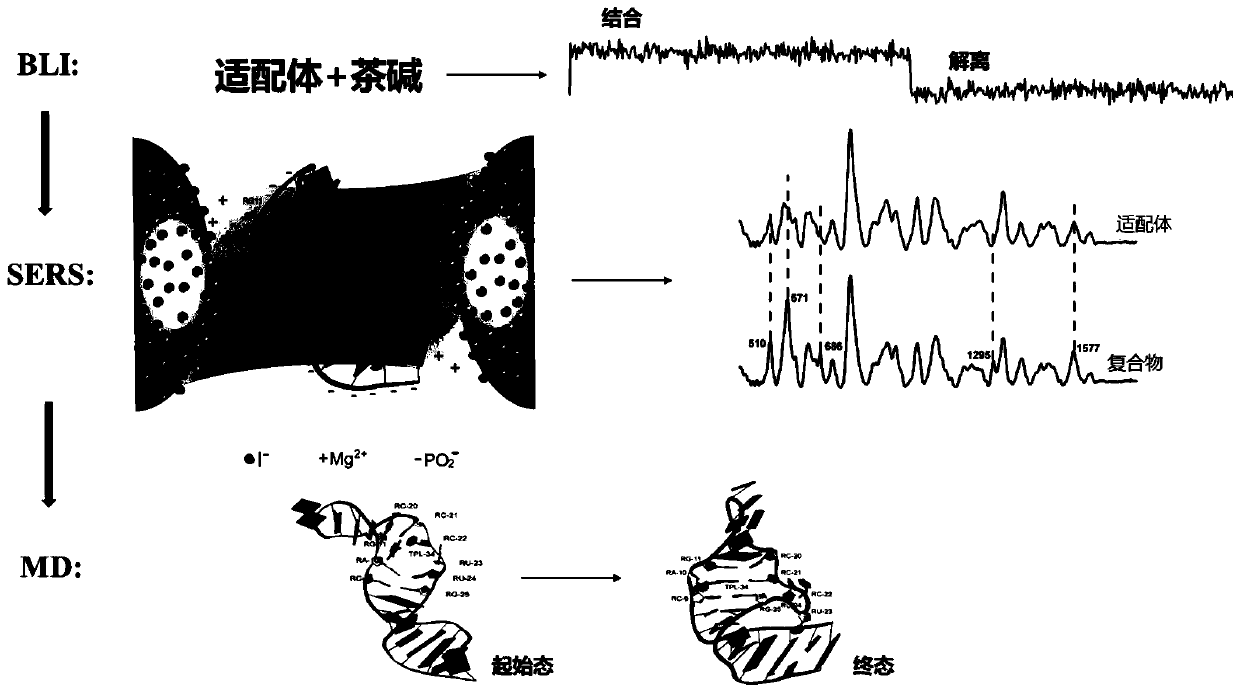
Method for rapidly and effectively detecting conformational change in aptamer and ligand small molecule binding process
A technology of combining process and aptamer, applied in the field of biomedicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
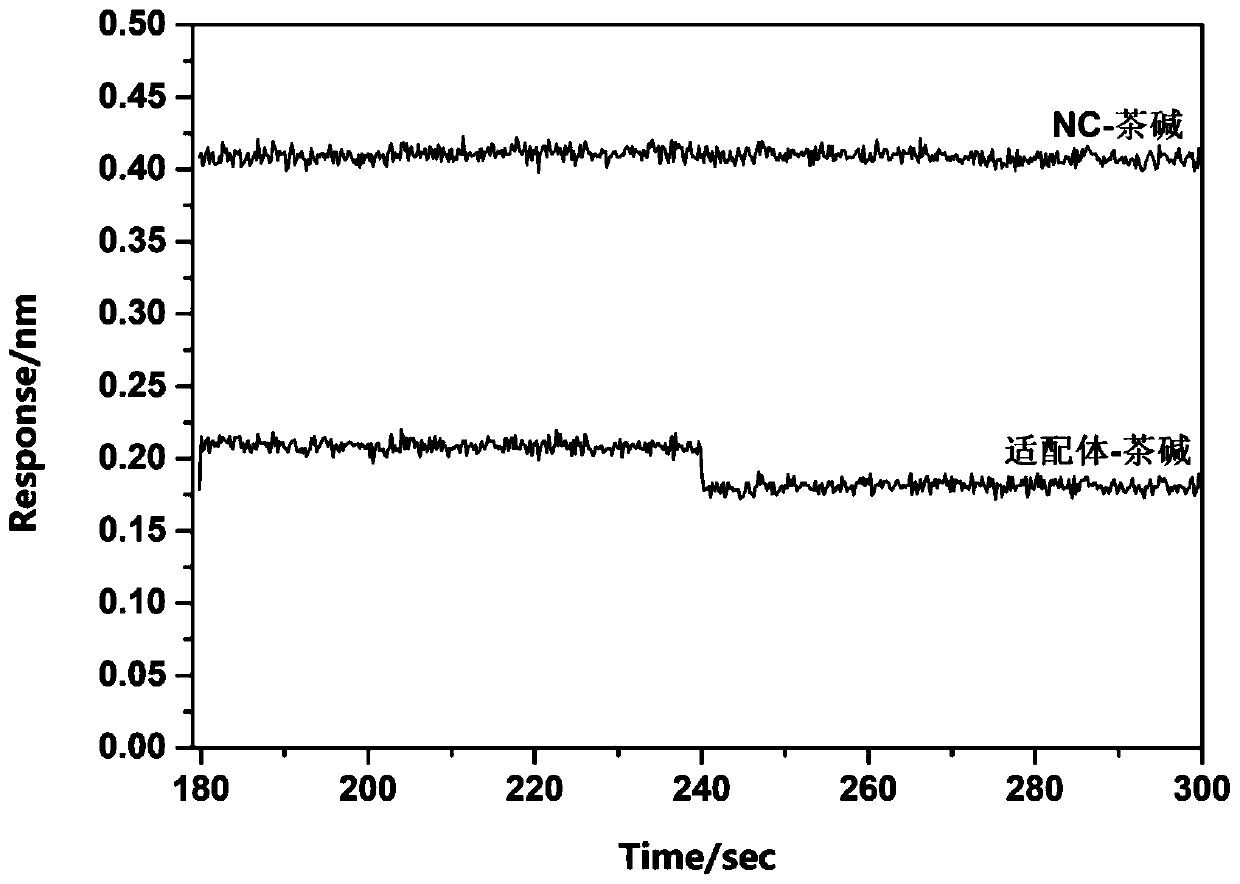
[0049] Embodiment 1: BLI measures the affinity between the small molecule theophylline and its aptamer RNA
[0050] The affinity between theophylline small molecule and its aptamer was determined by BLI technology, and at the same time, a 21-base RNA sequence (NC: UUGUACUACACAAAAAGUACUG, SEQ ID NO.2) was randomly selected as a negative control and theophylline reaction, and determine its affinity constant K under the same conditions as the aptamer d value.
[0051] First, use buffer (Tris-HCl buffer+10mM MgSO 4 ,pH 7.4) to prepare 10μM theophylline solution, 500nM aptamer RNA (with biotin at the 5' end) solution and 500nM NC (with biotin at the 5' end) solution respectively, and all the RNA solutions should be used immediately Ready to serve, store in ice box to prevent degradation. Then add 200 μL each of prepared aptamer solution, NC solution, theophylline, and buffer solution into each reaction well, and then dip the chip connected with streptomycin into each reaction we...
Embodiment 2
[0052] Example 2: Weak binding of aptamer RNA to a SERS substrate
[0053] First by the Lee method (Lee, P.C.; Meisel, D.Adsorption and surface-enhanced Ramanof dyes on silver and gold sols. The Journal of Physical Chemistry 1982,86,3391-3395.) to prepare silver colloid solution, and the obtained silver The gel solution is concentrated about 60 times by centrifugation and removing the supernatant, so as to enhance the Raman signal of the substance to be tested. However, when the silver colloid is concentrated, the impurities on the surface of the silver nanoparticles (AgNPs) are also enriched, showing a strong Raman signal, even exceeding the signal of the substance to be tested. Therefore, we added 5 μL of 1 mM KI solution to 10 μL of the concentrated silver colloid solution to clean the citrate ions on the surface of the silver nanoparticles and make the surface of the silver nanoparticles highly negatively charged (AgNP-I - ). However, due to the presence of a large numbe...
Embodiment 3
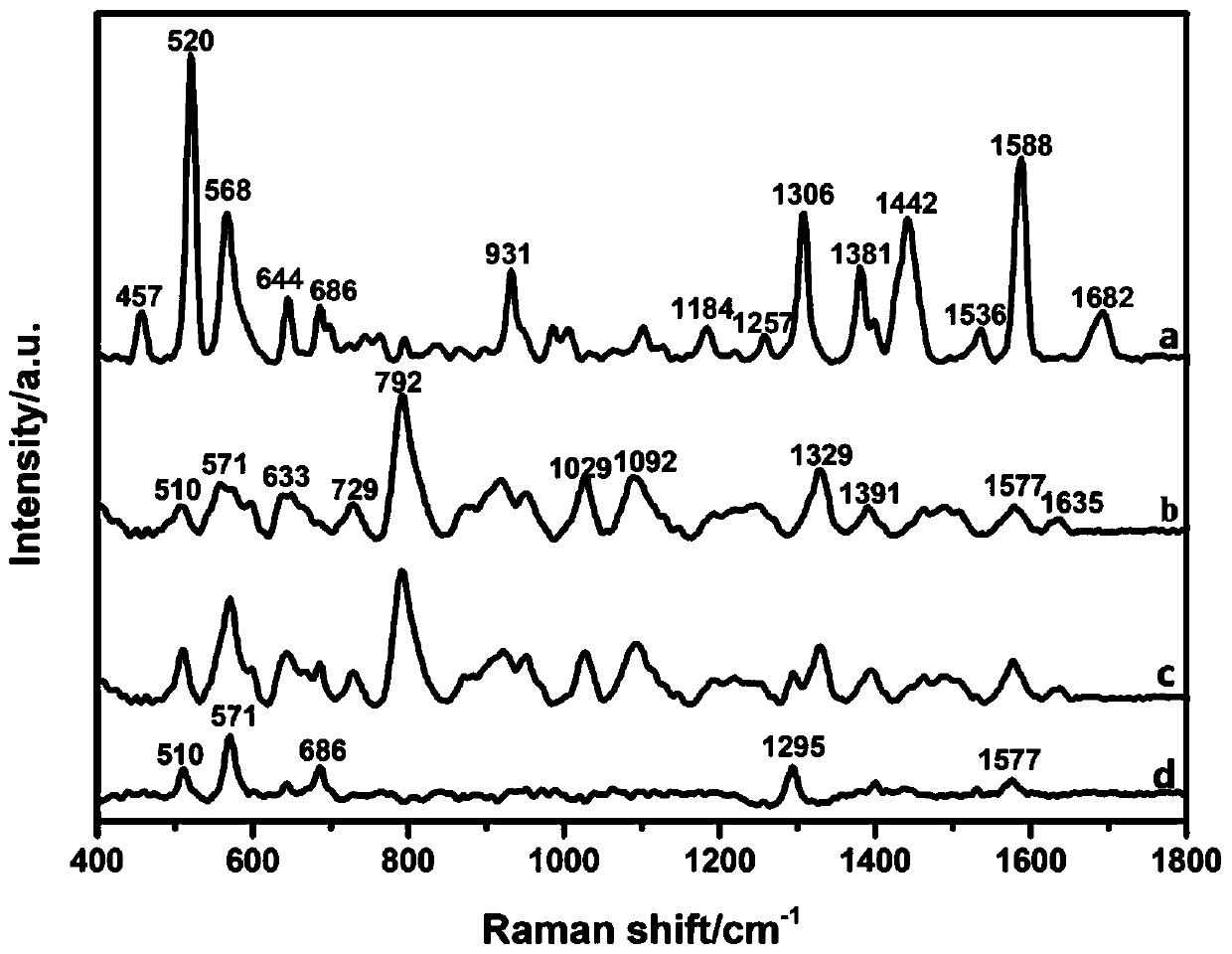
[0054] Example 3: Changes in the SERS spectra before and after the aptamer binds to theophylline
[0055] After the aptamers were weakly bound to the surface of the silver colloid, 100 μM theophylline solution was added in equal proportions. in Mg 2+ With the help of theophylline and the aptamer, theophylline and the aptamer are mutually induced to combine. Compared with the aptamer, the theophylline molecule has a smaller mass and volume, so in the process of the combination of the two, the conformation of theophylline molecule remains unchanged, while The aptamer continuously changes conformation to accommodate the entry of theophylline, and finally reaches a stable state. At this point, the mixed aptamer-theophylline solution was added to deionized water, diluted to 100 μL, and transferred to a 96-well plate to collect SERS signals. The final detection concentration of the substances to be tested was 10 μM, and the results were as follows: image 3 shown. After theophyl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com