High-throughput sequencing method of macrobenthos COI genes and application of high-throughput sequencing method of macrobenthos COI genes
A benthic, high-throughput technology, applied in the fields of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of low data accumulation, high-throughput, high error rate, avoid false positives, large classification The effect of learning resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] This embodiment provides a method for high-throughput sequencing of benthos COI gene, comprising the following steps:
[0054] First, the samples involved in this embodiment are prepared as follows: representative sampling points are set according to the characteristics of the water body such as geography and water quality. Use a Sauber net (30×30cm, net hole diameter 0.5mm) or a Peterson dredger (opening area 1 / 16m2) to collect samples of benthic animals in different habitats, collect 3 parallel samples at each point, and put them away after rough cleaning. Put it into a 300mL standard sample bottle and store it in 95% alcohol.
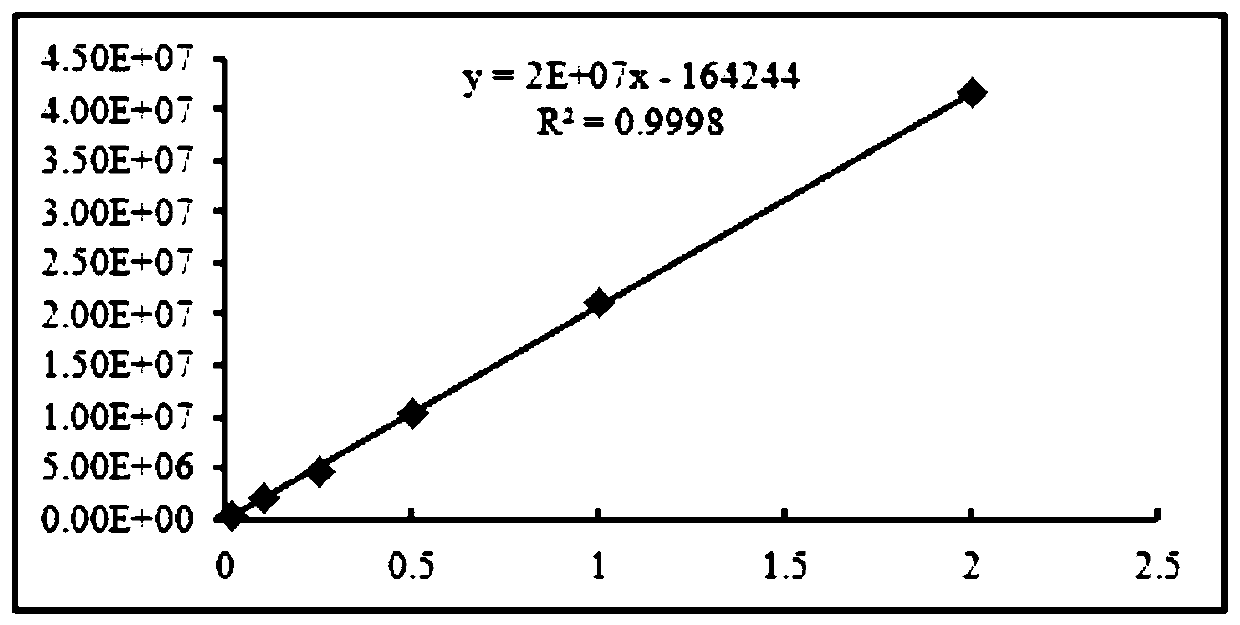
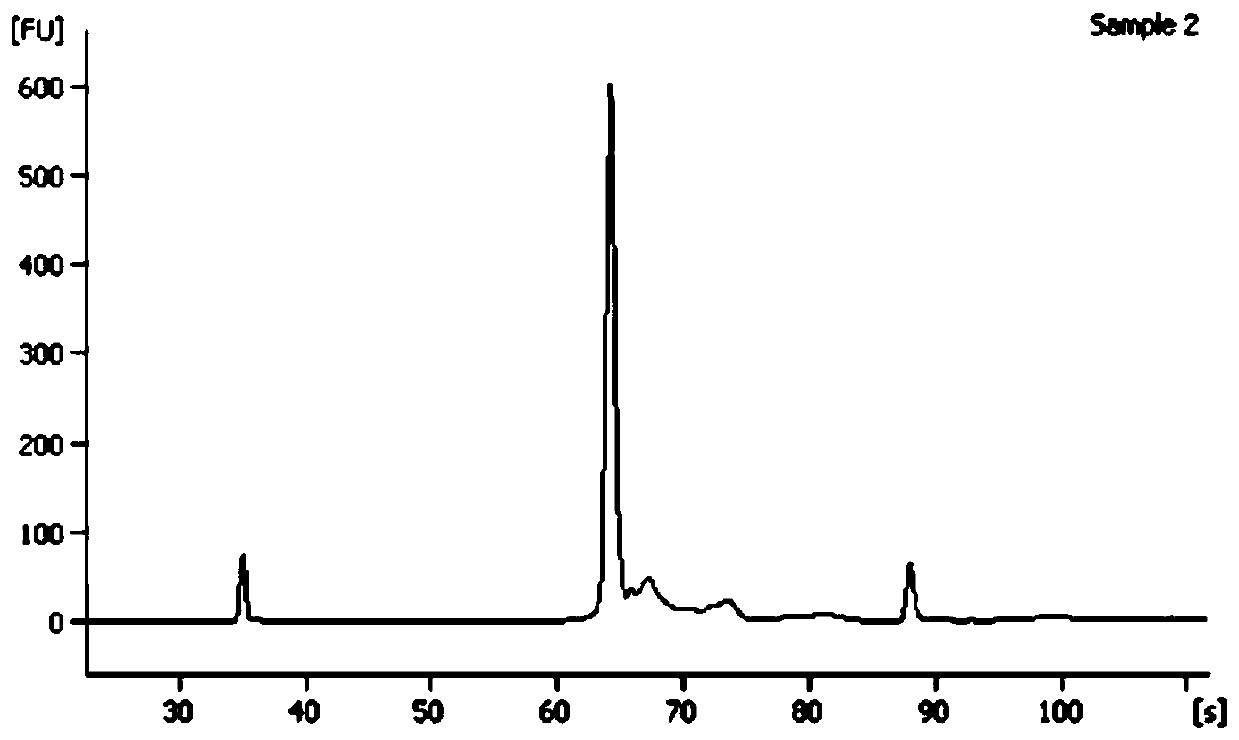
[0055] (1) DNA extraction was performed on the biological mixture sample prepared above using a tissue genomic DNA extraction kit, and the DNA concentration was measured and uniformly diluted to a final concentration of 5 ng / μl. The DNA with the same concentration after dilution is used as a template, the forward primer COIF is shown in SEQ I...
Embodiment 2
[0089] The sequencing results obtained in Example 1 are subjected to data analysis, including the following steps:
[0090] 1) Input the list file of MID sequences, run Split-libraries.py in QIIME software to classify the MIDs of different samples, remove degenerate bases, low quality scores, chimeras and noise sequences shorter than 150bp, and obtain Pure data.
[0091] 2) Use Usearch and CD-HIT software to classify OTUs of the pure data obtained above, and use multiple thresholds such as 97% and 98% for OTU picking to enhance the reliability of the results. The OTU representative sequence was matched with the sequence in the BOLD database with 80% sequence similarity for species annotation analysis, and the OTU list was generated.
[0092] 3) Calculate the α-diversity index of the sample based on the relative abundance of OTU (such as Shannon index, Simpson index), Chao index. Using the sequence abundance data and the appearance or absence data of OTUs to perform cluster a...
Embodiment 3
[0093] Embodiment 3 Water ecological detection application
[0094] Based on the data obtained in Example 2, the environmental stress factors and major pollutants affecting the macrobenthos community in the watershed were identified: the hydrochemical parameters were standardized by logarithmic transformation. By analyzing the differences in the number of taxa and environmental factors of various types of environmental factors in different groups, one-way analysis of variance is performed, and then the distance-based redundancy analysis dbRDA is carried out, and the invalid environmental variables are gradually deleted by using the pre-selection method (P>0.05), the Monte Carlo permutation test screened out the environmental stress factors and pollutants that affect the distribution of macrobenthos communities, and the relationship between community distribution and environmental factors was reflected through the ranking chart, and the macrobenthos that affected the watershed w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com