Kit for rapid screening CTG repetition of high-risk gene TCF4 for Fuchs endothelial corneal dystrophy
A technology of kits and repeated regions, applied in the field of molecular biology diagnosis, can solve the problems of increasing treatment complexity and side effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030]Example 1: Preparation of Fuchs corneal endothelial dystrophy rapid screening kit
[0031] 1. Primer design
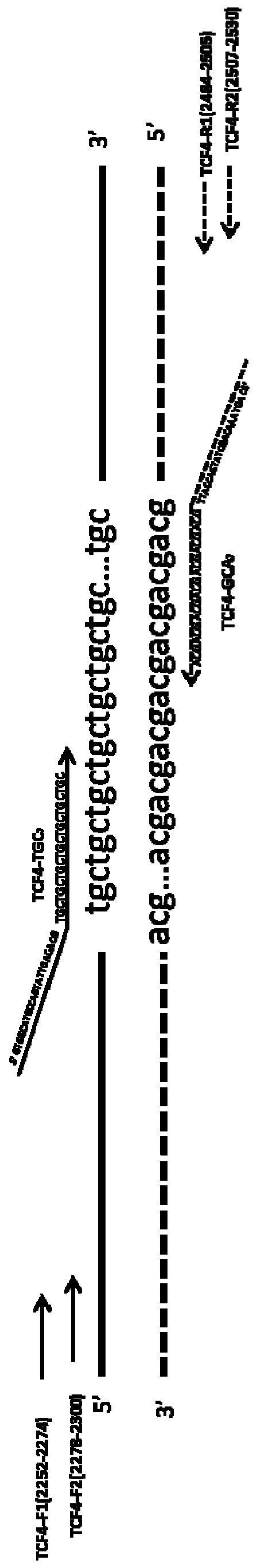
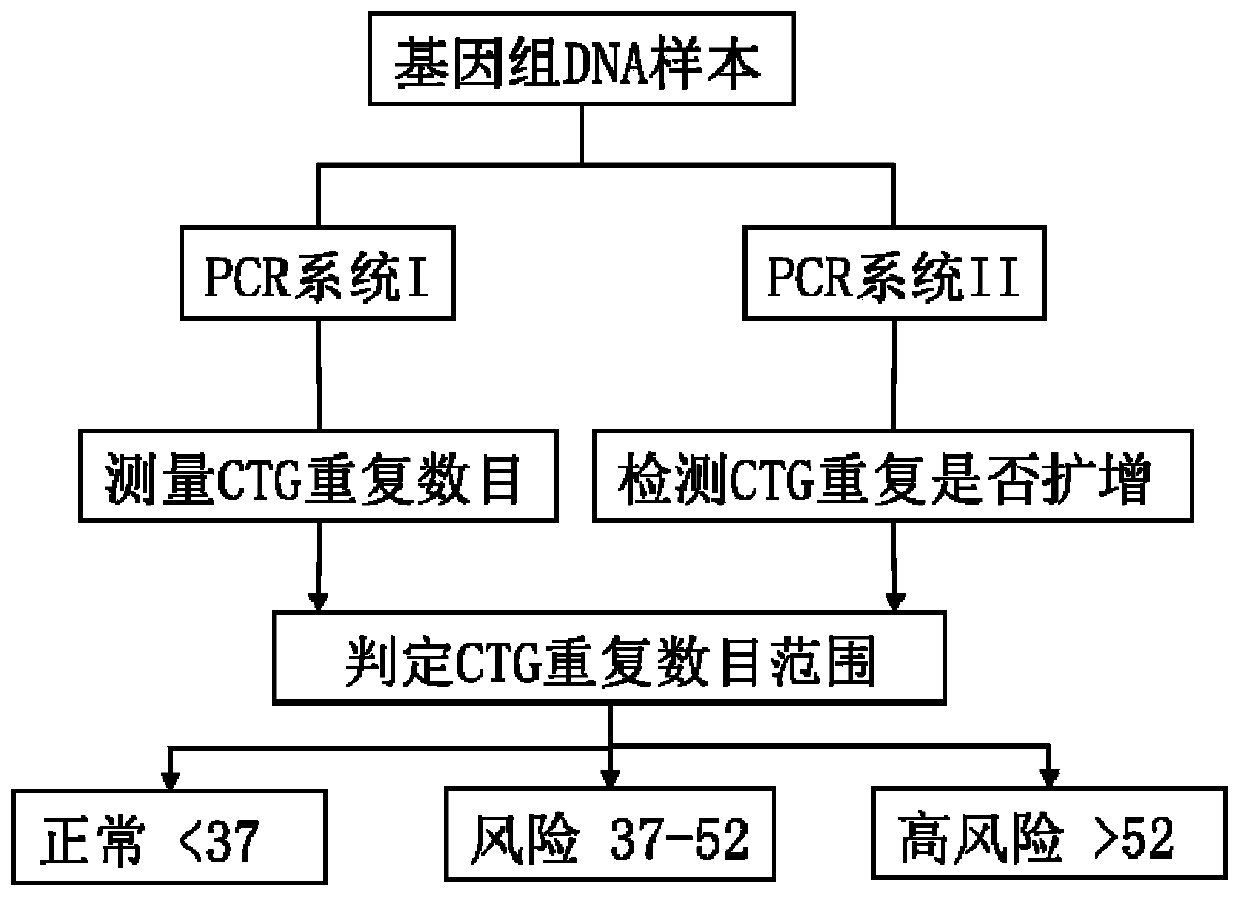
[0032] A pair of PCR System I specific upstream primer TCF4-F1 and downstream primer TCF4-R1 were designed within 200 bp of the upstream 5' end and downstream 3' end of the CTG repeat sequence of the TCF4 gene to amplify the target genome including the CTG repeat region, 5' flanking region and 3' flanking region. The 5' flanking region is 149bp, the 3' flanking region is 30bp, and the length of the amplified fragment is (231+3xn)bp, where n represents the number of CTG repeats. The formula for calculating the CTG repeat number is: CTG repeat number=(PCR product length-231) / 3.
[0033] In the CTG repeat region of the TCF4 gene, the upstream primer TCF4-TGC was designed to specifically bind to the CTG repeat sequence 7 , and added human genome irrelevant sequence GTGGCATGCCAGTATTGAGACG at its 5' end, primer TCF4-T1 sequence is the added sequence GTGGCATGCCAGTATT...
Embodiment 2
[0040] Example 2: Normal CTG Repeat
[0041] Use a commercially available genomic DNA extraction kit to extract genomic DNA from the peripheral blood of the sample, with a concentration of at least 50 ng / μL, a 260 / 230 value greater than 2.0, and a 260 / 280 value greater than 1.8.
[0042] PCR system I reaction system prepared using the kit in Example 1: prepare 20 μL PCR system in a 200 μL PCR thin-walled tube.
[0043] The PCR reaction system is as follows:
[0044] 10×PCR buffer 2μL MgCl 2 (50mmol / L)
2μL dNTP (10mmol / L) 0.4μL TCF4-F1 (10μmol / L) 1μL TCF4-R1 (10μmol / L) 1μL DNA polymerase 0.1μL dna 1μL 2×PCR enhancer 10μL double distilled water Make up to 20 μL
[0045] The sequences of the primers TCF4-F1 and TCF4-R1 are respectively: SEQ ID NO: 1 and SEQ ID NO: 2.
[0046] PCR amplification program: 95°C pre-denaturation for 10 minutes; 95°C for 45 seconds → 57°C for 45 seconds → 72°C for 5 minutes, ...
Embodiment 3
[0049] Example 3: CTG repeat expansion
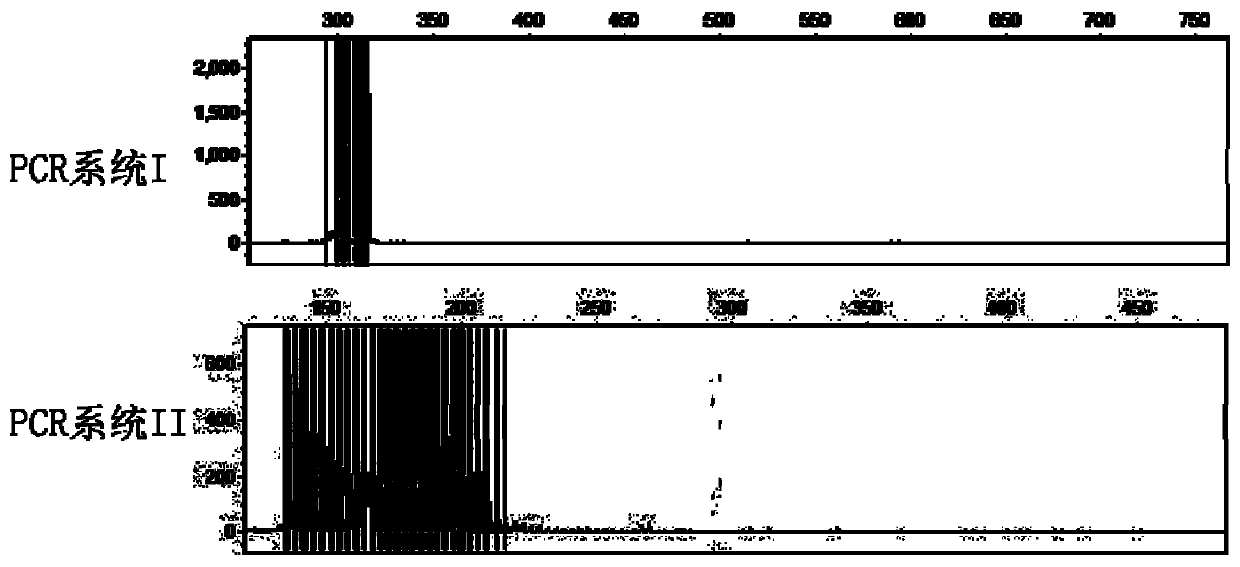
[0050] Use the kit in Example 1 to prepare PCR system I and PCR system II reaction systems: prepare 20 μL PCR system in a 200 μL PCR thin-walled tube.
[0051] The PCR reaction system is as follows:
[0052]
[0053] The sequence of primer TCF4-F2 is: SEQ ID NO: 6; the sequence of TCF4-R2 is: SEQ ID NO: 5; TCF4-TGC 7 The sequence is SEQ ID NO: 4; TCF4-GCA 7 The sequence is SEQ ID NO: 8; the sequence of TCF4-T1 is SEQ ID NO: 3; the sequence of TCF4-T2 is SEQ ID NO: 7.
[0054] PCR amplification program: 95°C pre-denaturation for 10 minutes; 97°C for 45 seconds→57°C for 45 seconds→72°C for 8 minutes, 40 cycles; 72°C for 10 minutes.
[0055] After the reaction, take 1 μL of PCR product, mix with 10 μL deionized formamide and 0.5 μL DNA internal standard 500LIZSize Standard, denature at 100°C for 5 minutes, and cool on ice. The mixture was analyzed for fragment length of PCR products with ABI 3500 Genetic Analyzer, the injection volt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com