Non-antibody-dependent protein methylation modification enrichment analysis method
An analytical method and methylation technology, applied in analytical materials, material separation, measurement devices, etc., can solve the problems of difficult to achieve deep coverage analysis of protein methylation modification, low enrichment efficiency of methylated modified peptides, etc. Achieve high-throughput analysis, improve selectivity, and reduce interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
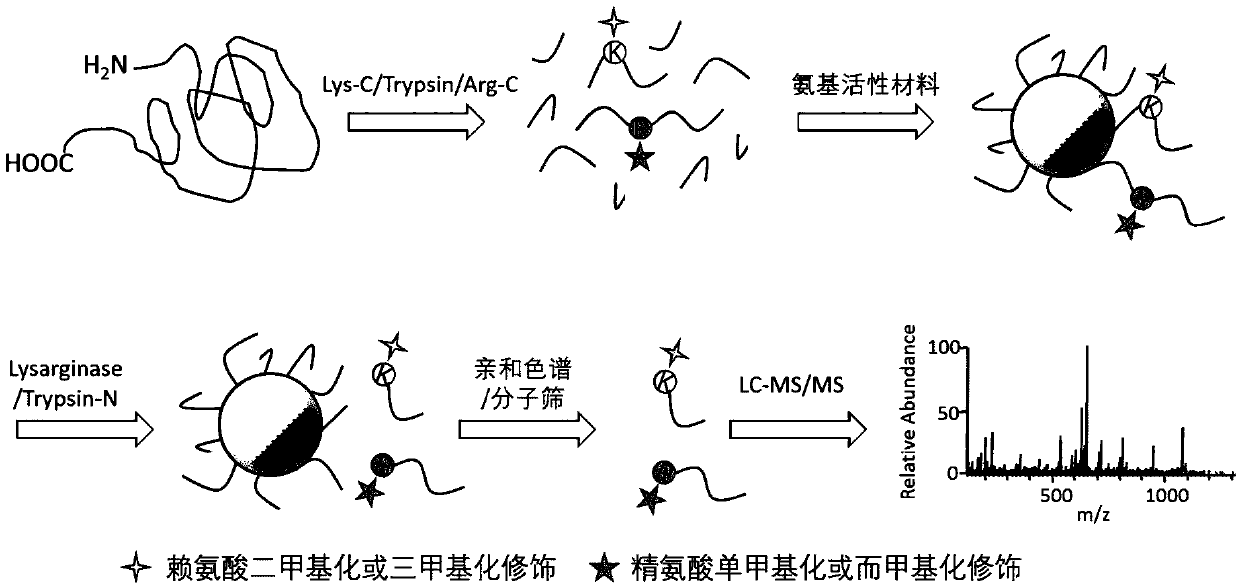
[0025] like figure 1As shown, after the protein is reductively alkylated, the protein is digested with Lys-C / Trypsin / Arg-C three enzymes, and then the peptides obtained by enzyme digestion are immobilized with amino active materials, and further used The enzyme Lysarginase or Trypsin-N with enzyme-cutting methylated modified lysine and arginine can digest the immobilized peptide, and use affinity chromatography or molecular sieve to separate the digested methylated peptide from the material , so as to realize the enrichment of methylated modified peptides, and analyze the enriched methylated modified samples by high performance liquid phase reverse chromatography-ultra-high resolution mass spectrometry (LC-MS / MS).
[0026] Using cervical cancer Hela cells as samples, 8M urea was used as the lysate for protein extraction, the extracted protein was quantified by BCA, and 1 mg of protein was taken for analysis of methylation modification. Add dithiothreitol to a final concentrat...
Embodiment 2
[0028] Using cervical cancer Hela cells as samples, protein was extracted using 6M guanidine hydrochloride as lysate, the extracted protein was quantified by BCA, and 1 mg of protein was taken for analysis of methylation modification. Add dithiothreitol to a final concentration of 10mM, after denaturation and reduction at 56°C for 30min, add iodoacetamide to a final concentration of 30mM, react at room temperature in the dark for 30min, then add dithiothreitol to the reaction system to a final concentration of 30mM, incubate at room temperature for 30min to terminate excess iodoacetamide. Add 10 μg of Lys-C, adjust the pH to 7.4, digest at 37°C for 4 hours, then dilute guanidine hydrochloride to 0.8M with 50 mM ammonium bicarbonate solution, add 20 μg of Trypsin, and digest at 37°C for 12 hours. Centrifuge at 15,000 rcf for 20 minutes to obtain enzyme-cleaved peptides, and immobilize the centrifuged peptide samples on a C18 reversed-phase trapping column (4.6mm i.d.×1cm) for d...
Embodiment 3
[0030] Take cervical cancer Hela cells as samples, use 6M guanidine hydrochloride as the lysate to extract protein, after the extracted protein is reductively alkylated, add 10 μg of Lys-C, adjust the pH to 7.4, and digest at 37 degrees for 4 hours , and then use 50 mM ammonium bicarbonate solution to dilute urea to 0.8 M, add 40 μg of Trypsin, and digest at 37° C. for 6 h. Centrifuge at 15,000 rcf for 20 minutes to obtain enzyme-cleaved peptides, and immobilize the centrifuged peptide samples on a C18 reversed-phase trapping column (4.6mm i.d.×1cm) for desalting, and use 0.1% trifluoroacetic acid / 80% acetonitrile for elution, After freeze drying. Then the dried peptide was dissolved in 20mM HEPES containing 5mM calcium chloride, 5mM dithiothreitol, and 2mM EDTA, and the pH was adjusted to 7.4, and 10μg of Arg-C was added to adjust the pH to 7.4 , Digest overnight at 37 degrees. Use C18 to desalt and dry the sample, add 5mg of HPG-ALD and 20mM sodium cyanoborocyanide to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com