Susceptible fungal gene LrWRKY-S1 of lily and application thereof
A technology of lrwrky-s1 and fungi, which is applied in the fields of application, genetic engineering, plant genetic improvement, etc., and achieves the effect of good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Cloning and Sequence Analysis of LrWRKY-S1 Gene of Lilium Minjiang
[0025] Using the leaves of Lily of the Minjiang River as the material, using TRIzol TM Plus RNA Purification Kit (12183555, Invitrogen TM ), extract total RNA according to the instructions, use DNase I (18047019, InvitrogenTM) to remove residual trace DNA, and use a spectrophotometer to measure the concentration of RNA for future use.
[0026] About 2.0 μg of total RNA from leaves of Lilium Minjiang was taken, and the first-strand cDNA was synthesized according to the instructions of PrimeScript II first-strand cDNA synthesis kit (6210A, Takara).
[0027] PCR amplification system: 2xfast pfu master Max 10ul, forward primer (LrWRKY-S1-1-F, 10μM) 1μL, reverse primer (LrWRKY-S1-R, 10μM) 1μL, template (cDNA) 1μL, sterile ddH 2 O to make up to 20 μL.
[0028] The forward and reverse primers are:
[0029] LrWRKY-S1-F:5'-GTCCGAATATCATGGACGGAG-3'
[0030] LrWRKY-S1-R:5'-CTACAACATTTAAACGAAGAAGGCA...
Embodiment 2
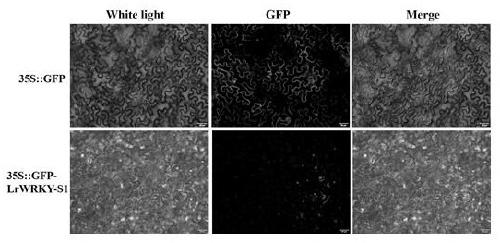
[0035] Example 2 Construction of GFP fusion expression vector and analysis of subcellular localization
[0036] (1) Construction of pTF101-P35S::GFP-LrWRKY-S1 expression vector
[0037] According to the pTF101-GFP vector sequence and the full-length sequence of the LrWRKY-S1 gene (SEQ ID NO.1), a forward primer (LrWRKY-S1-inf-F1) and a reverse primer (LrWRKY-S1-inf-R1) were designed. Using the TA-ligated positive clone plasmid in Example 1 as a template, PCR amplification of the LrWRKY-S1 gene fragment was performed.
[0038] The primer sequences are as follows:
[0039] LrWRKY-S1-inf-F1:
[0040] 5'- ATGGACGGAGGCTCTGGGACAG-3'
[0041] LrWRKY-S1-inf-R1:
[0042] 5' TTAAACGAAGAAGGCAGAGGCATCG-3'
[0043] The underline is the In-fusion cloning vector linker sequence, and the bold sequence is the restriction site.
[0044] PCR reaction system: high-fidelity amplification enzyme PrimeSTARHS (R010 A, TaKaRa) 0.5μL, 5xPrimeSTARBuffer (Mg 2+ Plus) 10 μL, forward primer (10 ...
Embodiment 3
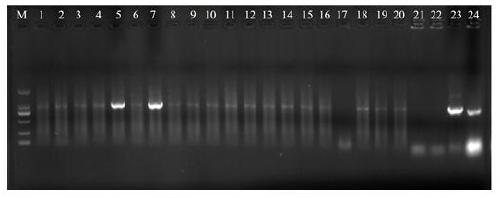
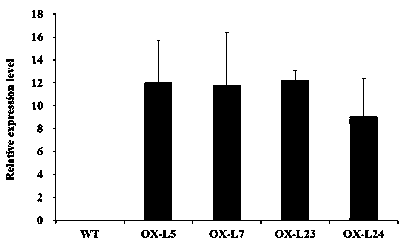
[0054] Example 3 LrWRKY-S1 Gene Overexpression Vector Construction and Genetic Transformation of Arabidopsis
[0055] (1) Construction of pBI121-P35S::LrWRKY-S1 overexpression vector
[0056] According to the pBI121 vector sequence and LrWRKY-S1 gene sequence (SEQ ID NO.1), design primers LrWRKY-S1-inf-F2 and LrWRKY-S1-inf-R2, and introduce seamless cloning (In-fusion) vector adapter sequence into the primers and enzyme cleavage site sequences. The specific amplification of the LrWRKY-S1 gene fragment was performed using the TA-ligated positive clone plasmid in Example 1 as a template.
[0057] The primer sequences are as follows:
[0058] LrWRKY-S1-inf-F2:
[0059] 5'- GGACTCTAGAGGATCC GTCCGAATATCATGGACGGAG-3'
[0060]LrWRKY-S1-inf-R2:
[0061] 5'- GATCGGGGAAATTCGAGCTC CTACAACATTTAAACGAAGAAGGCAG-3'
[0062] Among them, the underline is the linker sequence of the In-fusion cloning vector.
[0063] The PCR reaction system is: high-fidelity amplification enzyme PrimeS...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com