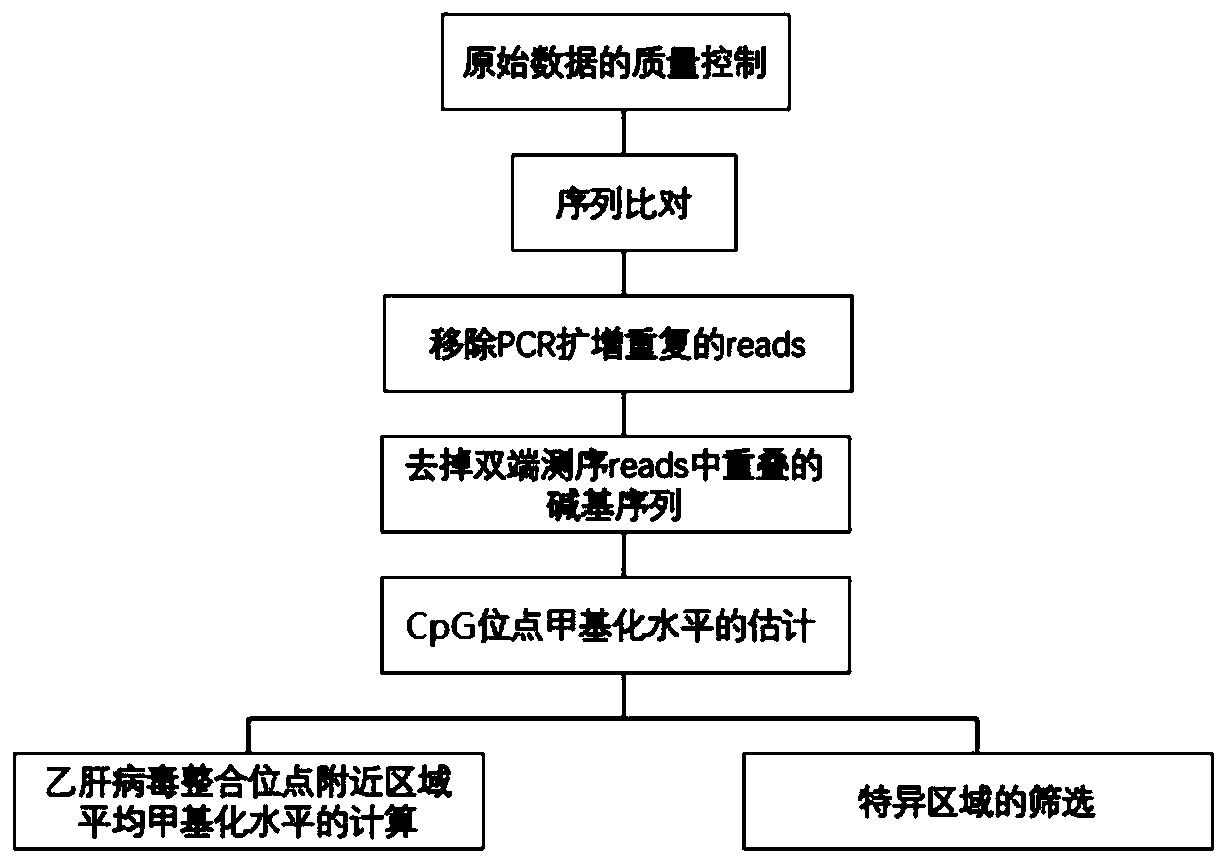
Application of methylation status near HBV integration site in cancer detection
An integration site, methylation technology, applied in the field of molecular biology, can solve the problems of low tumor DNA, unable to achieve the performance of research samples, and limited large-scale application.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
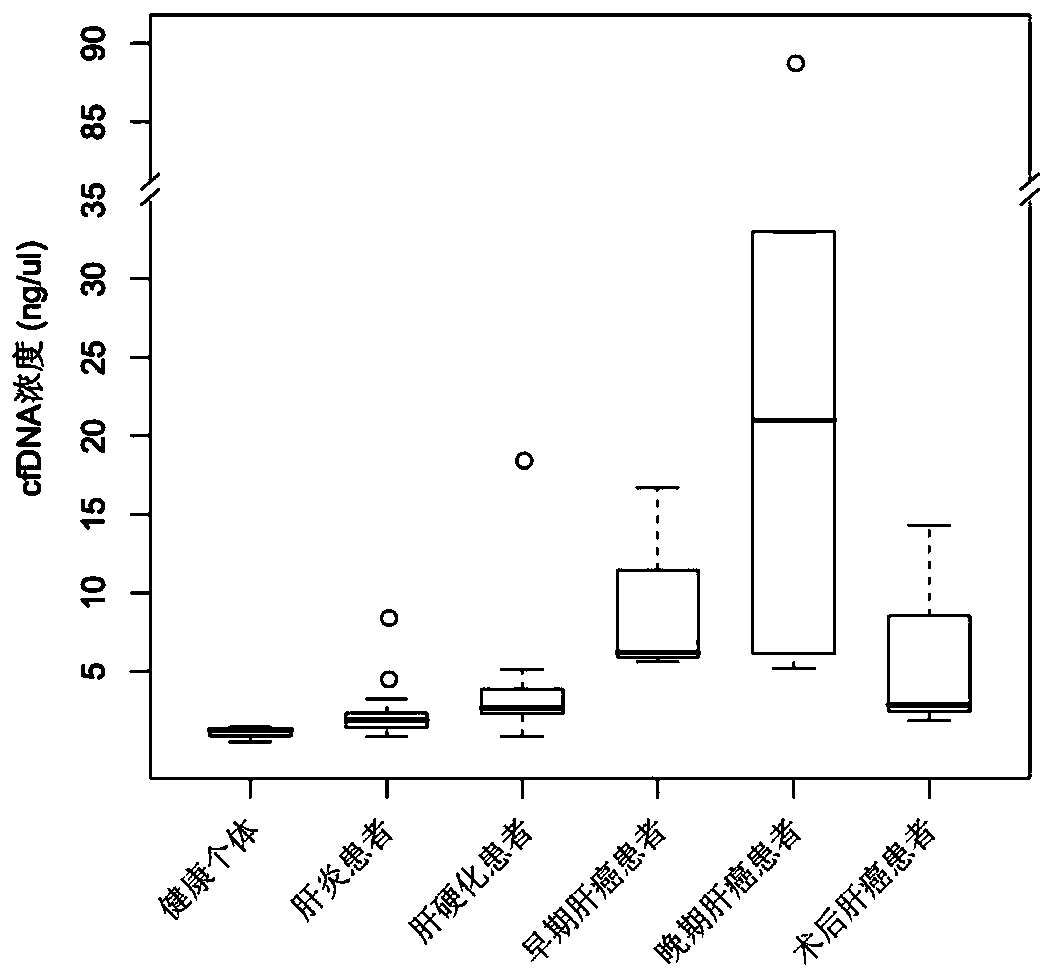
[0117] Samples from 54 subjects were collected, including 17 patients with liver cancer (3 patients with early liver cancer, 5 patients with advanced liver cancer, and 9 patients after surgery for liver cancer), 17 patients with liver cirrhosis, 17 patients with hepatitis and 3 healthy patients Individual peripheral blood samples. The clinical information of the above subjects was collected, including age, sex, hepatitis B virus (Heptitis B Virus, HBV) infection status, tumor size, alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (Tbil ) and alpha-fetoprotein (AFP), etc. According to the Barcelona Liver Cancer Clinical Staging System (BCLC), patients with liver cancer were divided into early and late stages, with stages A and B considered early stages, and stages C and D considered advanced stages.
[0118] Take 10ml of peripheral blood in a Streck Cell-Free DNA BCT anticoagulant tube, centrifuge at 3,000×g for 15 minutes at 4°C, absorb the su...
Embodiment 2
[0138] The specific process of performing WGBS sequencing on cfDNA samples is as follows:
[0139] (1) Add A for end repair: Use the Bioo Scientific Kit kit to absorb 10ng of cfDNA sample, add λ-5mc as an internal reference at a ratio of 5‰, and use ddH 2 O make up the volume to 32μl (ie DNA Mix and water). Configure a 50 μL reaction system as follows:
[0140]
[0141] The reaction conditions were set as follows: 22°C for 20 minutes, 72°C for 20 minutes, and kept at 4°C, and placed in a PCR instrument.
[0142] (2) Repaired samples plus methylated adapters (NEXTflex TM Bisulfite-Seq Adapters): Dilute adapters to 3 μM (14 μL H 2 (0+2 μL stock solution); add 47.5 μL of Ligase Enzyme Mix to the repaired 50 μL reaction solution, then add 2.5 μL of the diluted linker, and react at 22°C for 15 minutes.
[0143] (3) Magnetic bead purification:
[0144] 1) Take out the AMPure magnetic beads from the 4°C refrigerator half an hour in advance and place them at room temperature ...
Embodiment 3
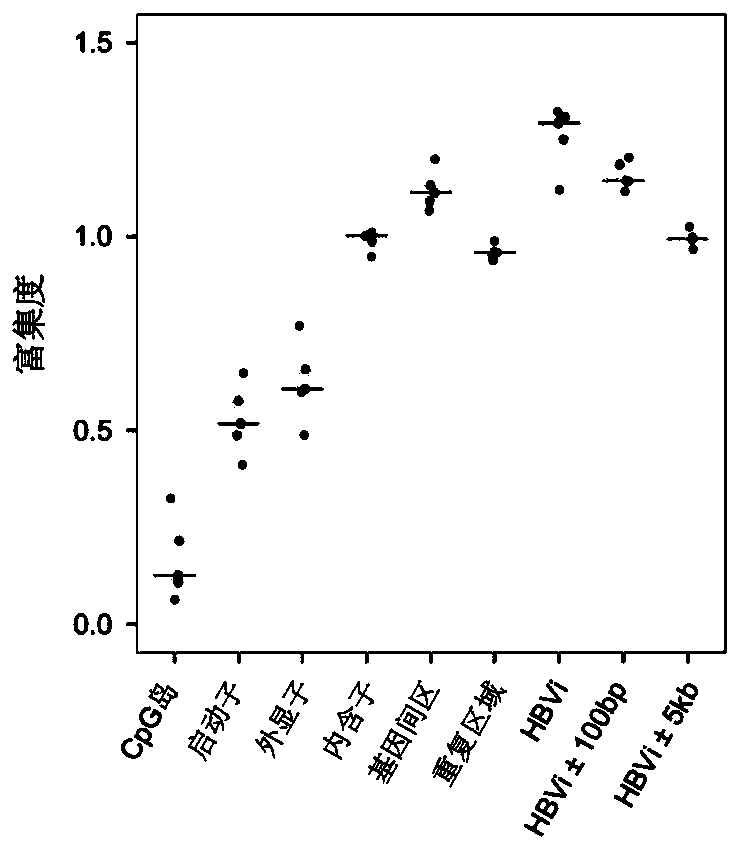
[0173] The inventors have discovered an unexpected phenomenon, in free DNA samples, CpG sites tend to be enriched in introns, intergenic regions, repeat (repeat) regions and regions near the HBV integration site (HBVi), especially The HBV integration site and its upstream and downstream vicinity, but the CpG sites in the CpG island region are very rare. In order to show and illustrate this phenomenon, in the present embodiment, from the sample of embodiment 1, extract healthy individual, chronic hepatitis, liver cirrhosis, advanced liver cancer, postoperative liver cancer patient each one case, carry out the WGBS of general depth to total free DNA ( The average sequencing depth is 58M sequencing read pairs, and other conditions are the same as in Example 2), and the enrichment of CpG sites is counted, and the results are as follows image 3 shown. Among them, the highest enrichment degree was reached at the HBV integration site, and the enrichment degree of CpG sites in the u...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com