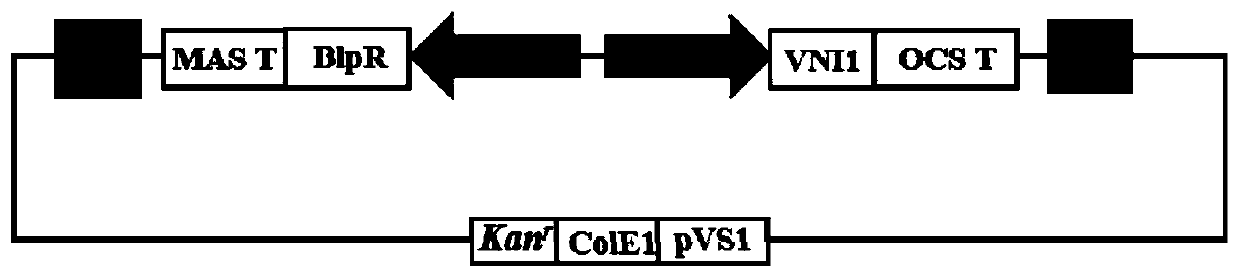
Plant salt-resistant protein MsVNI1 as well as coding gene and application thereof
A plant and protein technology, applied in the field of plant genetic engineering, can solve the problems of improving plant yield and quality molecular design, insufficient plant salt-tolerant gene resource bank, etc., to achieve the effect of increasing leaf size and salt-tolerant quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Cloning of MsVNI1 gene
[0032] The primers: MsVNI1-F and MsVNI1-R were designed on both sides of the full-length sequence of MtVNI1, a species of Medicago truncatula with high homology in the genome, and PCR amplification was performed using the cDNA of Medicago sativa as a template.
[0033] The primer sequences are as follows:
[0034] MsVNI1-F: CCCTTCCGTGCTTCATA
[0035] MsVNI1-R: CTCAAATAAATCAGCCATCA
[0036] The PCR reaction system is: 2 μL cDNA, 5 μL 10×Buffer, 4 μL 2.5mMdNTP, 10 μM forward / reverse primers 1 μL each, 0.5 μL 5U / μL Taq enzyme and 36.5 μL ddH 2 O. Mix well after adding the sample on ice. The PCR reaction conditions were: 94°C for 5 min; 94°C for 30 sec, 56°C for 45 sec; 72°C for 2 min, 34 cycles; 72°C for 10 min.
[0037] PCR amplified product was carried out 1% agarose gel electrophoresis detection, obtained the fragment that fragment size is 1544bp ( figure 1 ), carried out gel recovery (using Promega gel recovery kit) to the a...
Embodiment 2
[0038] Example 2: Construction of recombinant vector and transient expression in tobacco cells to observe subcellular localization
[0039] Using the sequence fragment obtained above as a template, MsVNI1 was designed with a linker primer infused with the expression vector pCABIA1300-cGFP, and the fragment was amplified with high-fidelity enzymes; the expression vector pCABIA1300-cGFP was digested with restriction endonuclease BamHI. The MsVNI1 gene fragment and the pCABIA1300-cGFP vector fragment were recovered. The two recovered fragments were then connected by homologous recombination using Infusion enzyme (purchased from Vazyme). The ligation product was transformed into Escherichia coli DH5α competent cells by heat shock method, added 600 μL of liquid LB medium, incubated for 1 h, spread on LB plates of Khanna antibiotics, and incubated at 37°C for 14 h. Single-clonal colonies were picked, amplified and cultured in liquid LB medium containing kanamycin, and subjected to ...
Embodiment 3
[0041] Embodiment 3: Obtaining of overexpressing MsVNI1 transgenic Arabidopsis plants
[0042] The overexpression vector was designed to connect the primers of the entry vector: MsVNI1-OE-F and MsVNI1-OE-R, and the end of the primer was introduced into the BamHI restriction site and the 9 bases after the entry vector pENTRE restriction site (Infusion linker sequence). The obtained full-length sequence of MsVNI1 was used as a template, and PCR amplification was performed with the above-mentioned primers.
[0043] The primer sequences are as follows:
[0044] MsVNI1-OE-F: TCCTTCACCCGGGATCCATGGCCGAAACAAAATTGAT
[0045] MsVNI1-OE-R: ACCCTTTATCGGGATCCCCAGCCATCATGTTTAGTCT
[0046] Among them, the underline is the Infusion linker sequence.
[0047] The above-mentioned amplified fragment is recovered. The pENTRE vector was digested with restriction endonuclease BamHI and recovered. The above two fragments were ligated and transformed into Escherichia coli to obtain a positive clo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com