Escherichia coli and application thereof to synthesis of fucosylated oligosaccharide
A technology of fucosyl and Escherichia coli, which is applied in the field of metabolic engineering, can solve the problems of unreachable industrial production, low yield of human milk oligosaccharides, and low yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
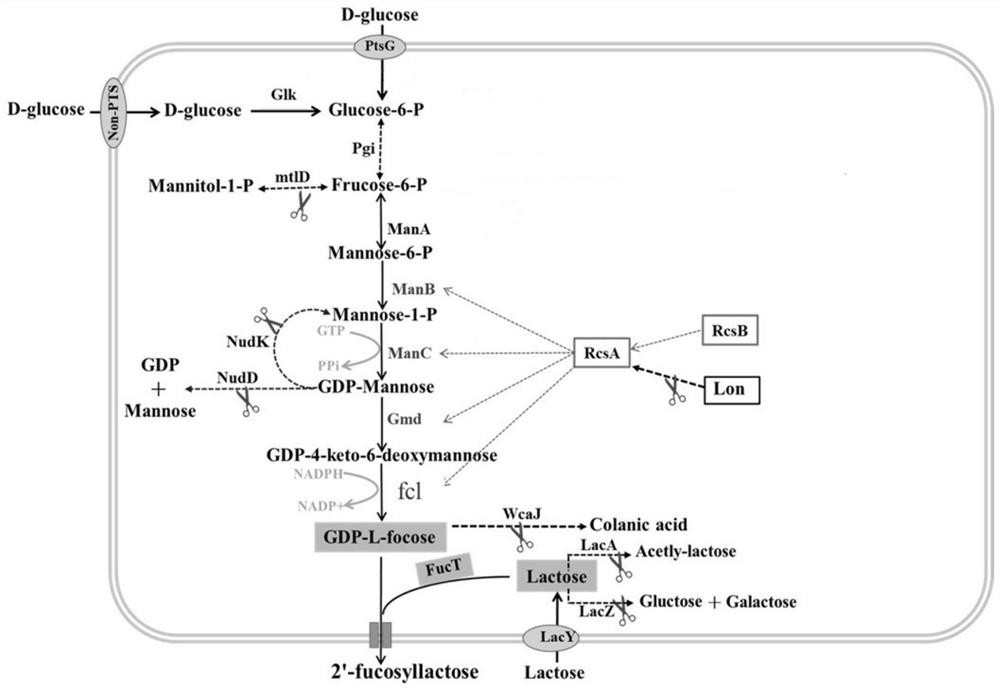
[0062] The research on the efficient synthesis of fucosylated oligosaccharides using engineered Escherichia coli has been carried out for many years. Based on the existing technology, most of them use the metabolic intermediate fructose-6-phosphate of Escherichia coli as the starting point for the de novo synthesis of GDP-L-fucose way.
[0063] For the construction method of metabolic engineering bacteria for efficiently biosynthesizing fucosylated human milk oligosaccharides using sucrose in this example, see figure 1 , including the following steps:
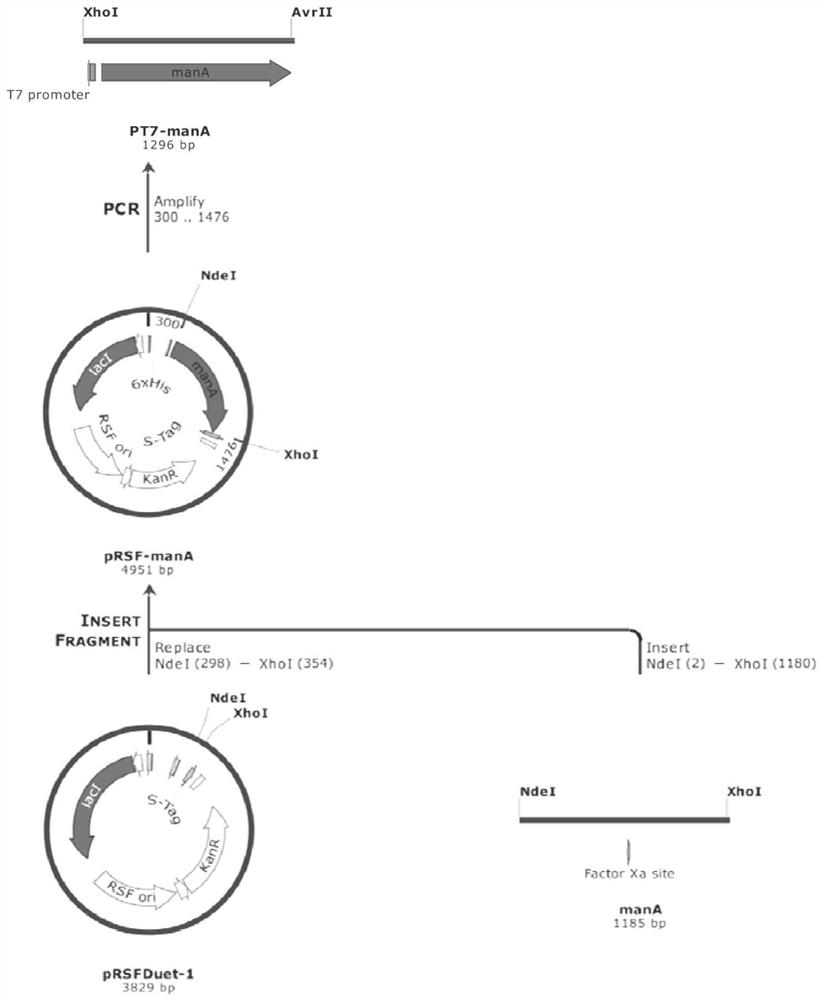
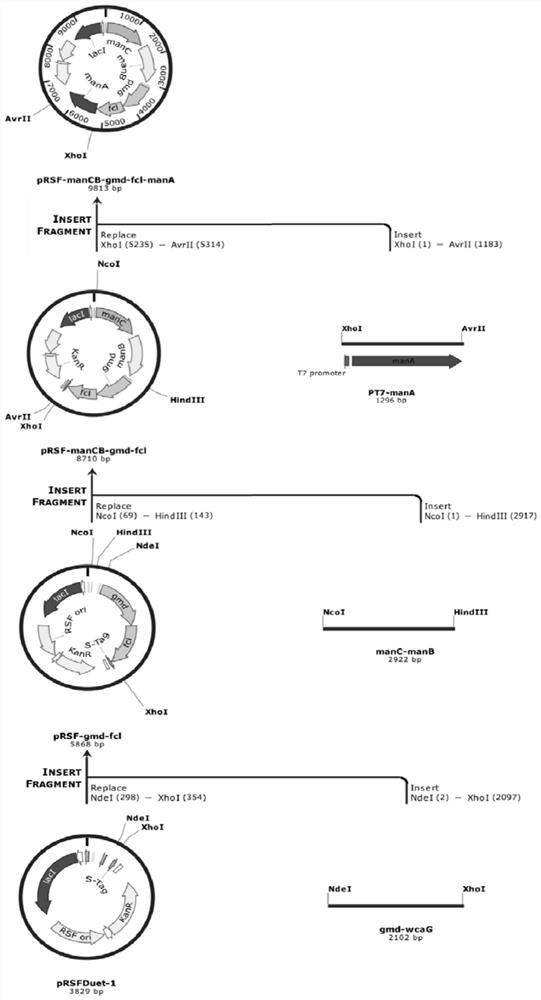
[0064] (1) Construct engineering strains and overexpress the de novo synthesis pathway genes of GDP-L-fucose. Such as image 3 with Figure 4As shown, the 6-phosphomannose isomerase gene manA (Gene ID: 944840) and the phosphomannose mutase manB derived from E. coli str.K-12substr.MG1655 (GenBank: NC_000913.3) were amplified by PCR respectively. (Gene ID: 946574), α-D-mannose 1-phosphate guanyltransferase manC (Gene ID: 946...
Embodiment 2
[0082] Example 2 Using Escherichia coli to ferment and synthesize 2'-fucosyl lactose (5L tank)
[0083] (1) Seed medium LB (g / L): tryptone 10, yeast extract 5, sodium chloride 10, pH7.2-7.4; when preparing solid medium, add 17g / L agar powder;
[0084] Initial fermentation medium (g / L): glucose 10-20, ammonium sulfate 3-7, dipotassium hydrogen phosphate 8-12, potassium dihydrogen phosphate 6-10, citric acid 0.5-1.0, CaCl 2 1.0~1.5, vitamin B10.01~0.1, defoamer 20~80ml / L, trace element mother solution 10~20mL / L; trace element mother solution formula (g / L): nitrilotriacetic acid (add appropriate amount of alkali, separate Preparation) 8~12, ferric ammonium citrate 5~7, zinc sulfate heptahydrate 0.5~1, CoCl 2 ·6H 2 O 0.1~0.5, manganese chloride tetrahydrate 0.6~1.2, CuCl 2 2H 2 O 0.1~0.2, boric acid 0.1~0.5, Na 2 MoO 4 2H 2 O 0.1~0.5; feed solution (g / L): sucrose 500~800, magnesium sulfate 10~20.
[0085] (2) Inoculate the engineered Escherichia coli constructed in Example...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com