Construction method and application of cell strain for expressing NTNG1 gene
A cell line and gene technology, applied in the direction of genetically modified cells, applications, cells modified by introducing foreign genetic material, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] 1. qRT-PCR detection of NTNG1 expression
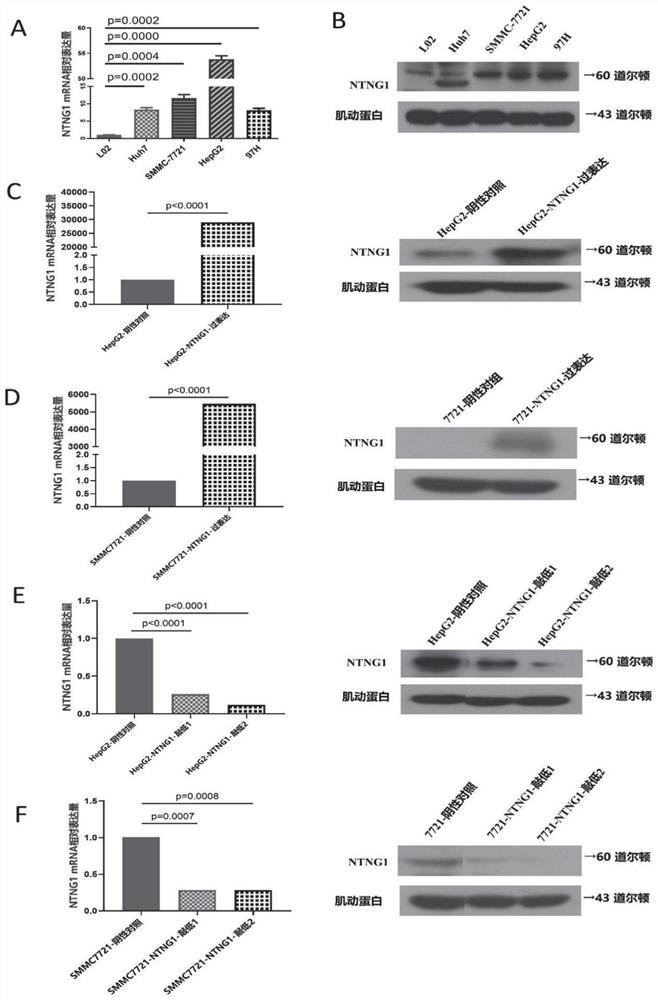
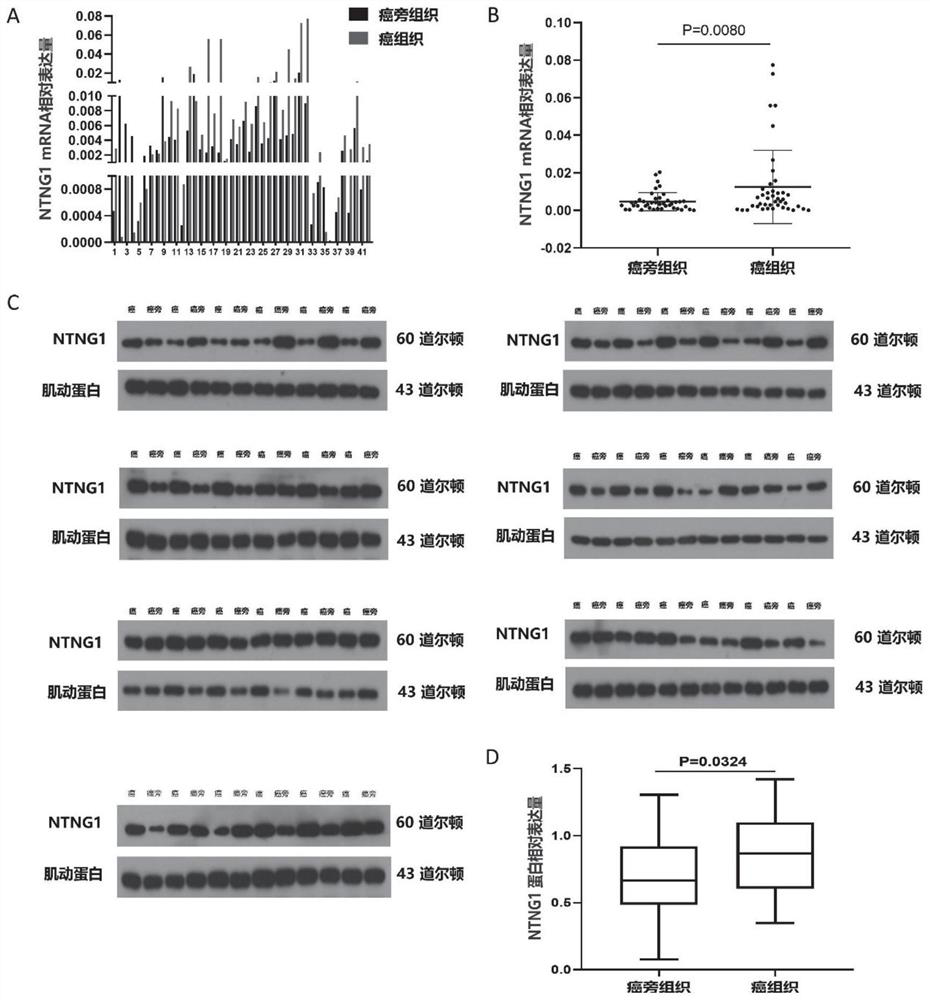
[0060] Fresh tissue specimens obtained during surgery were cooled in liquid nitrogen and stored in a -80°C freezer. After all the liver cancer tissues and paracancerous tissues were cut up, the total RNA was extracted with Trizol (TaKaRa, A-79061) reagent, and the total RNA was reverse-transcribed into cDNA according to the instructions of the kit. Reaction conditions: 95°C pre-denaturation, enzyme activation (30s), 95°C denaturation (5s), 60°C (30s), 40 cycles. With βactin as internal reference, with 2 -ΔΔCt The relative expression level of target gene NTNG1 mRNA was calculated by method. The upstream primer of NTNG1 is 5′-CTTGGAAGGAGTATCCCAAGC-3′, the downstream primer is 5′-TGTGGCATAATACTGATAGGGCT-3′; the upstream primer of β-actin is 5′-CACCATTGGCAATGAGCGGTTC-3′, and the downstream primer is 5′-AGGTCTTTGCGGATGTCCACGT-3′. The experiment was repeated three times. Conclusions such as figure 1 a.
[0061] Establishment of...
Embodiment 2
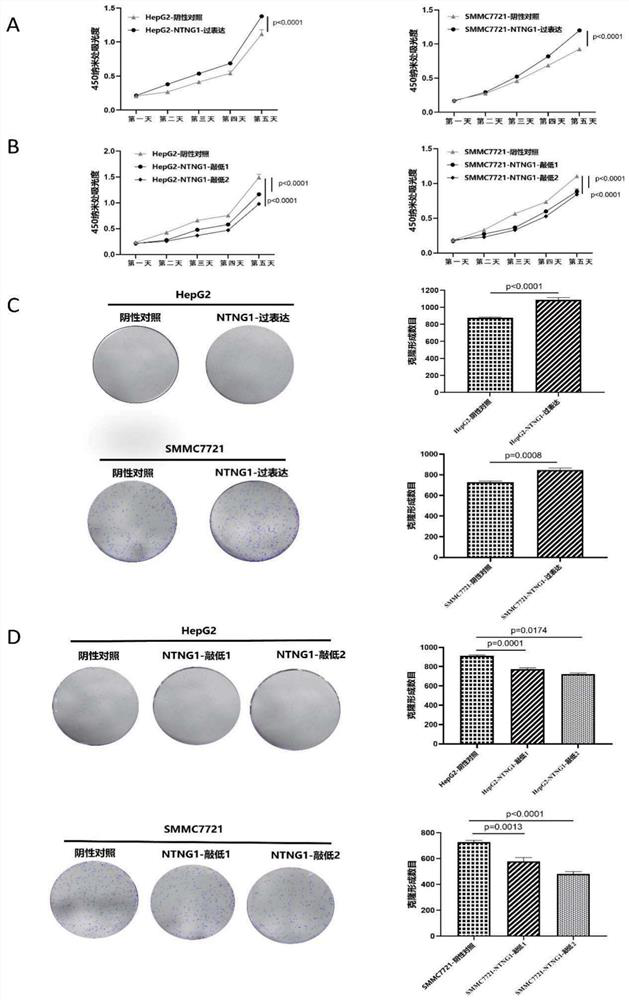
[0106] CCK-8 assay (cytotoxicity detection)
[0107] The specific steps of the CCK-8 experiment were performed according to the manufacturer's standard protocol. Cells were diluted in serum-free medium and seeded in 96-well cell culture plates at a density of 2000 cells / well. The plate was incubated at 37°C in 5% CO 2 incubator. To measure the growth rate of the cells, replace 100 μL of the spent medium with an equal volume of fresh medium containing 10% CCK-8. Cells were then further incubated at 37°C for 3 hours. Finally, a microplate reader (5082Grodig, Tecan, Austria) was used to measure the value of cell absorbance at 450 nm. The experiment was repeated 3 times and the results were recorded.
[0108] 1. Plate colony formation experiment
[0109] Cells in the logarithmic growth phase were taken from the cells in good growth state, digested with trypsin, resuspended with medium, inoculated into six-well plates with 500 cells per well, and cultured normally for 1520 da...
Embodiment 3
[0118] Flow Cytometry
[0119] Collect the HepG2 and SMMC7721 cells of each group transfected for 48 hours, digest with EDTA-free trypsin, wash twice with PBS, add 500 μL of binding buffer to resuspend the cells, and then add 5 μL of annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI), protected from light, incubated at room temperature for 15 min, and then detected the apoptosis rate of cells in each group on a flow cytometer (BD, USA). Then the cells were collected and fixed with 70% ethanol at 4°C for 2 hours, treated with PI (50 μg / mL) and RNaseA I (100 μg / mL), incubated at 37°C in the dark for 30 minutes, and placed in a flow cytometer The changes of DNA content in cells were analyzed in each period. Experiments were repeated three times.
[0120] Conclusion: NTNG1 inhibits apoptosis and regulates cell cycle progression in human hepatocellular carcinoma cells
[0121] The uncontrolled proliferation of tumor cells caused by apoptosis and cycle regulati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com