Rapid quantitative detection method capable of specifically identifying survival cells of enterobacter sakazakii and primer thereof
A quantitative detection method, the technology of Enterobacter sakazakii, is applied in the field of microbiology to achieve the effects of excellent detection accuracy, rapid quantitative detection, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
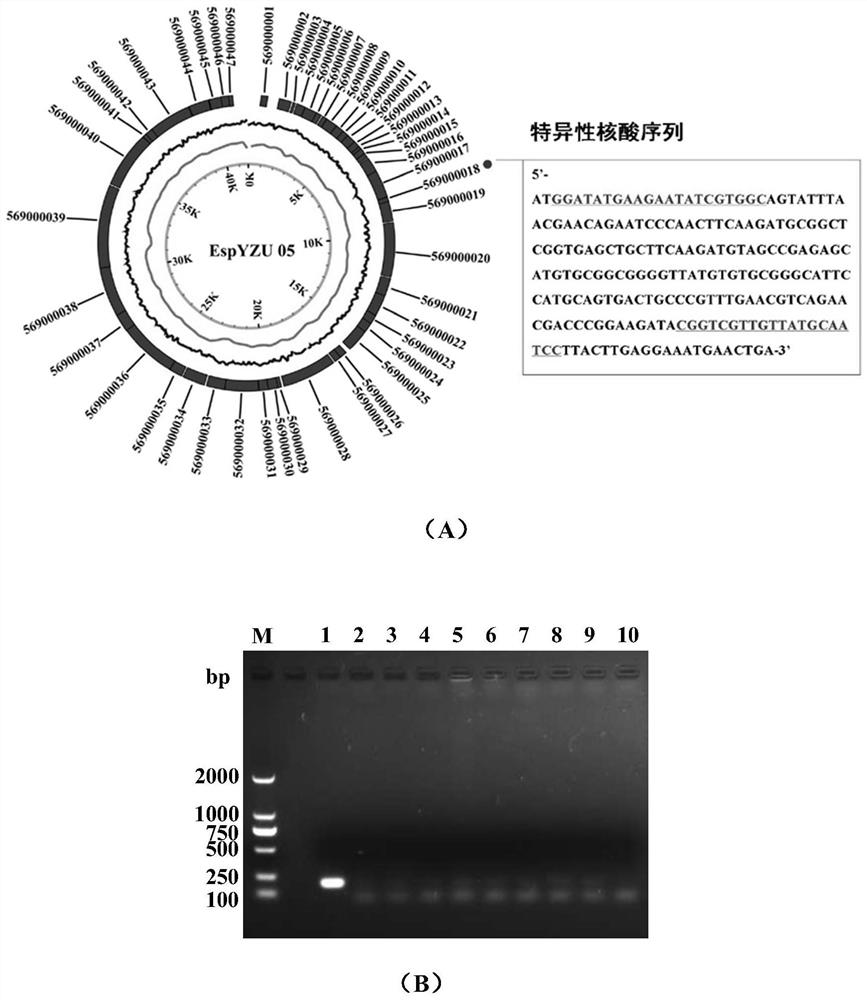
[0041] The design and verification results of specific primers for phage EspYZU05
[0042] Genomic DNA of phage EspYZU05 was extracted, the whole genome sequence was obtained by Illumina Hiseq sequencing, and the reading frame 569000018 (ORF18) with the lowest homology was selected as the specific nucleic acid detection target (sequence such as figure 1 Shown in A), the ORF18 nucleic acid sequence is SEQ ID NO.1: ATG GAT ATG AAG AAT ATC GTG GCA GTATTT AAC GAA CAG AAT CCC AAC TTC AAG ATG CGG CTC GGT GAG CTG CTT CAA GAT GTAGCC GAG AGC ATG TGC GGC GGG GTT ATG TGT GCG GGC ATT CCA TGC AGT GAC TGC CCGTTT GAA CGT CAG AAC GAC CCG GAA GAT ACG GTC GTT GTT ATG CAA TCC TTA CTT GAGGAA ATG AAC TGA; 29.1%. According to the upstream and downstream sequences of ORF18, the PCR upstream primer sequence is SEQ ID NO.2:5'-GGATATGAAGAATATCGTGGC-3' (named EspYZU05-ORF18-F1), and the downstream primer sequence is SEQ ID NO.3:5'-GGATTGCATAACAACGACCG-3 '(named EspYZU05-ORF18-R1), the expected size of...
Embodiment 2
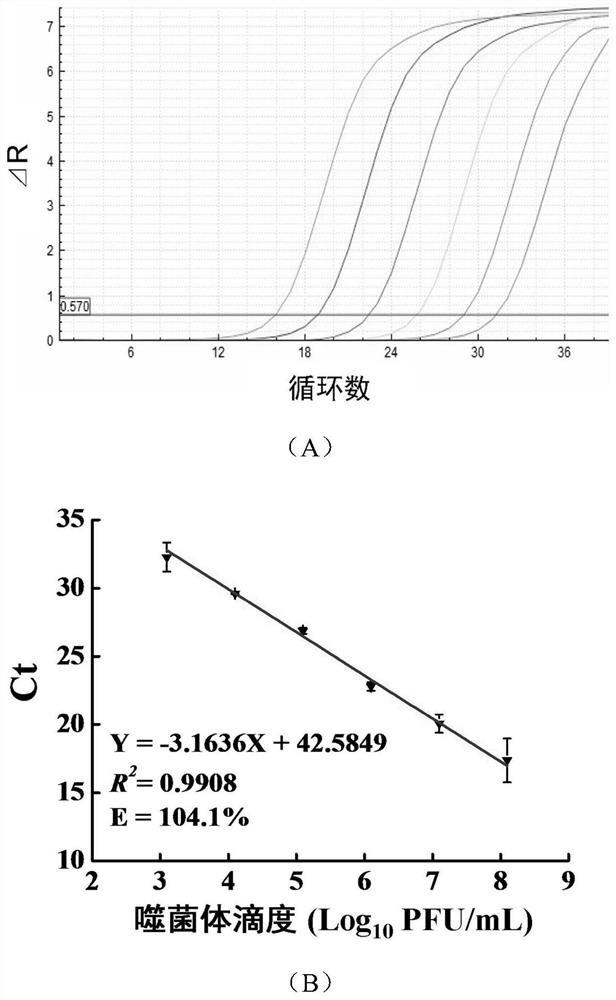
[0045] Establishment of qPCR Amplification System of Phage EspYZU05
[0046] The phage EspYZU05 (CCTCC NO: M 2016716) was mixed with Enterobacter sakazakii CICC 21596 according to the ratio of MOI = 1. After standing for 10 minutes, it was added to 10 mL of liquid medium, and cultured at 37°C and 150r / min for 8 hours. The culture solution was centrifuged at 6000r / min for 10min to take the supernatant, passed through a filter membrane with a pore size of 0.45μm, and the filtrate was stored at 4°C. The titer of phage in the filtrate was determined by the conventional double-layer plate method, and the titer of the phage suspension was adjusted to 10 with sterile SM buffer. 8 PFU / mL, spare. Take the phage EspYZU05 suspension and carry out 10-fold dilution to make 10 8 PFU / mL, 10 7 PFU / mL, 10 6 PFU / mL, 10 5 PFU / mL, 10 4 PFU / mL, 10 3 PFU / mL. Three replicates were performed for each dilution. For each dilution, 250 μL was taken in a boiling water bath for 5 min, and 2 μL wa...
Embodiment 3
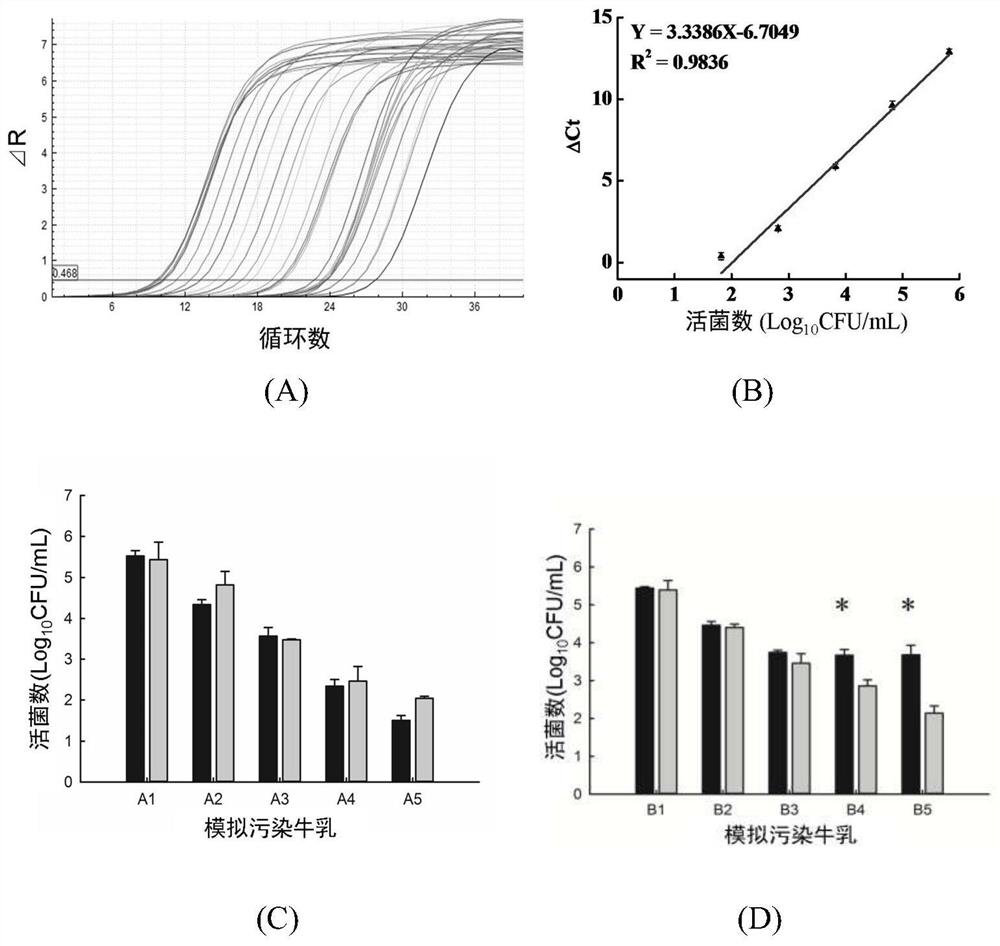
[0048] Rapid quantitative detection of Enterobacter sakazakii in simulated contaminated milk
[0049] Contains 10 3 Preparation of PFU / mL bacteriophage lysis medium: weigh 1g peptone, 0.3g beef extract, 0.5g NaCl, 0.5g CaCl 2 , 0.5g CaCl 2 , distilled water to 100mL, adjust the pH to 7.5, and sterilize at 121°C for 15min; after cooling, add filter-sterilized 10 3 The phage EspYZU05 suspension of PFU / mL, the formation final content is 10 3 PFU / mL of the lysed medium.
[0050] Preparation of simulated contaminated milk samples: add 12g skim milk powder to 100mL deionized water to make 12% skim milk powder, autoclave at 115°C for 20min, and set aside. Enterobacter sakazakii CICC21596 was added to aseptic skim milk with a mass fraction of 12%, homogenized and diluted to form a final concentration of 10 2 -10 6 Various sample gradient suspensions of CFU / mL were used as group A samples (sample numbers: A1-A5, 10 2 、10 3 、10 4 、10 5 、10 6 CFU / mL); Escherichia coli CICC1066...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com