Autophagy protein beclin-1 as well as coding gene and application thereof
A technology of beclin-1 and gene, applied in application, genetic engineering, plant genetic improvement, etc., can solve problems that threaten the sustainable development of industries, economic losses, and large-scale death of farmed oysters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Example 1: Cloning of full-length Beclin-1 gene of Hong Kong oyster
[0017] 1. RNA extraction
[0018] Total RNA was extracted using Trizol (Invitorgen), and the specific steps were as follows: (1) Take 50 mg of Hong Kong oyster tissue in a liquid nitrogen pre-cooled mortar, grind the tissue into powder, and transfer it to a centrifuge tube containing 1 ml Trizol , pipetting, and mixing; (2) Add 200 μL of chloroform, shake vigorously for 40 seconds, and place at room temperature for 5 minutes; (3) Centrifuge at 12,000×g for 10 minutes at 4°C, transfer the supernatant to a new tube; (4) Add 0.5ml of isopropyl (5) centrifuge at 12,000×g for 10 min at 4°C, discard the supernatant; (6) wash the precipitate twice with 75% ethanol; (7) discard the supernatant, add 50 μL DEPC water to dissolve the RNA .
[0019] 2. Cloning of Beclin-1 gene from Hong Kong oyster
[0020] According to the existing transcriptome data, the Beclin-1 partial sequence was screened, and the full-l...
Embodiment 2
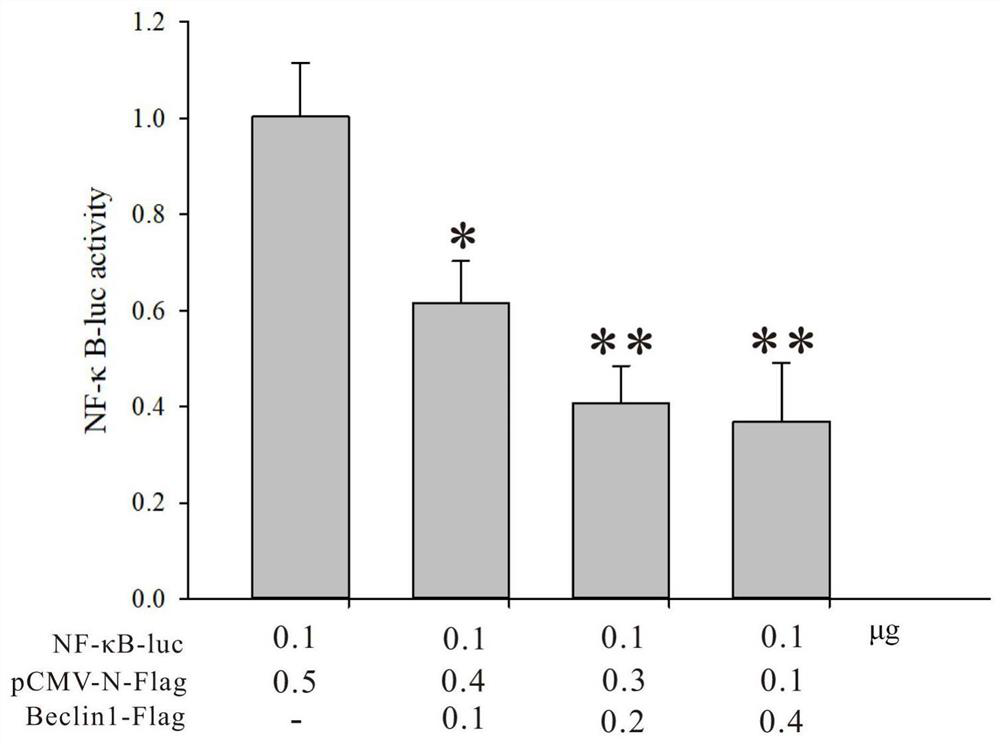
[0021] Example 2: Construction of eukaryotic expression vector of Hong Kong oyster Beclin-1 gene and luciferase experiment
[0022] 1. Construction of Hong Kong oyster Beclin-1 gene eukaryotic expression vector: (1) Design a pair of primers covering ChBeclin-1 ORF F1: CGTAGGATCCATGGCAACCATCAAGGTTGA; R1: GATCGTCGACTCACTTGTTGGCGAACTGAG, add BamH-I and SalI sites and protect base; (2) PCR amplification using the above primers; (3) PCR products and pCMV-N-Flag vectors were double-digested with BamH-I and SalI, and the purified enzyme-cleaved products were recovered; (4) ligase The cut PCR fragment and the pCMV-N-Flag vector; (5) transformation, picking positive clones, sequencing to verify that the vector is correct, and performing follow-up experiments to obtain Beclin1-Flag. The empty vector pCMV-N-Flag was used as a control.
[0023] 2. Cell Transfection and Luciferase Assay
[0024] HEK293 cells were seeded in 48-well plates (1×10 5 cells / well), liposomes were co-transfecte...
Embodiment 3
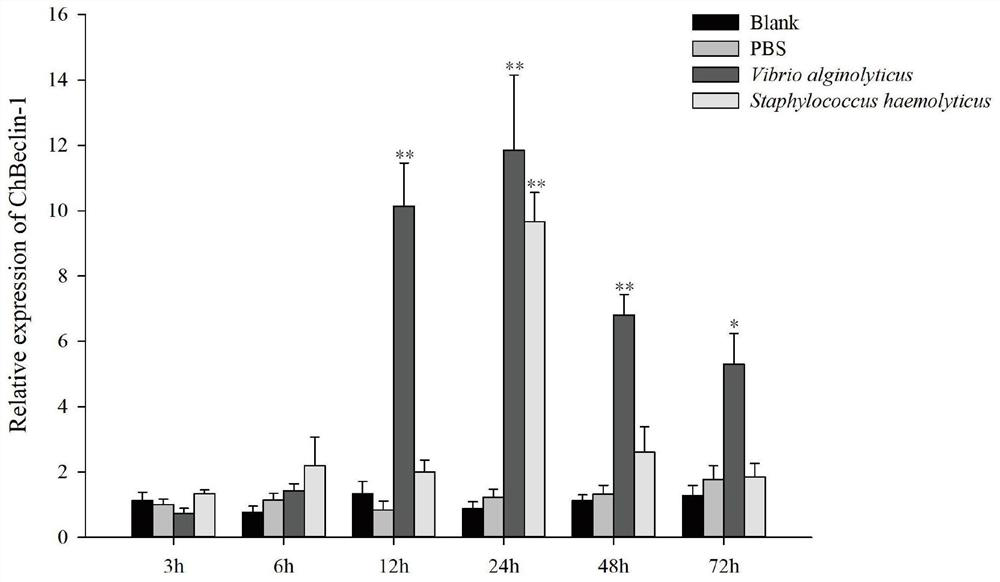
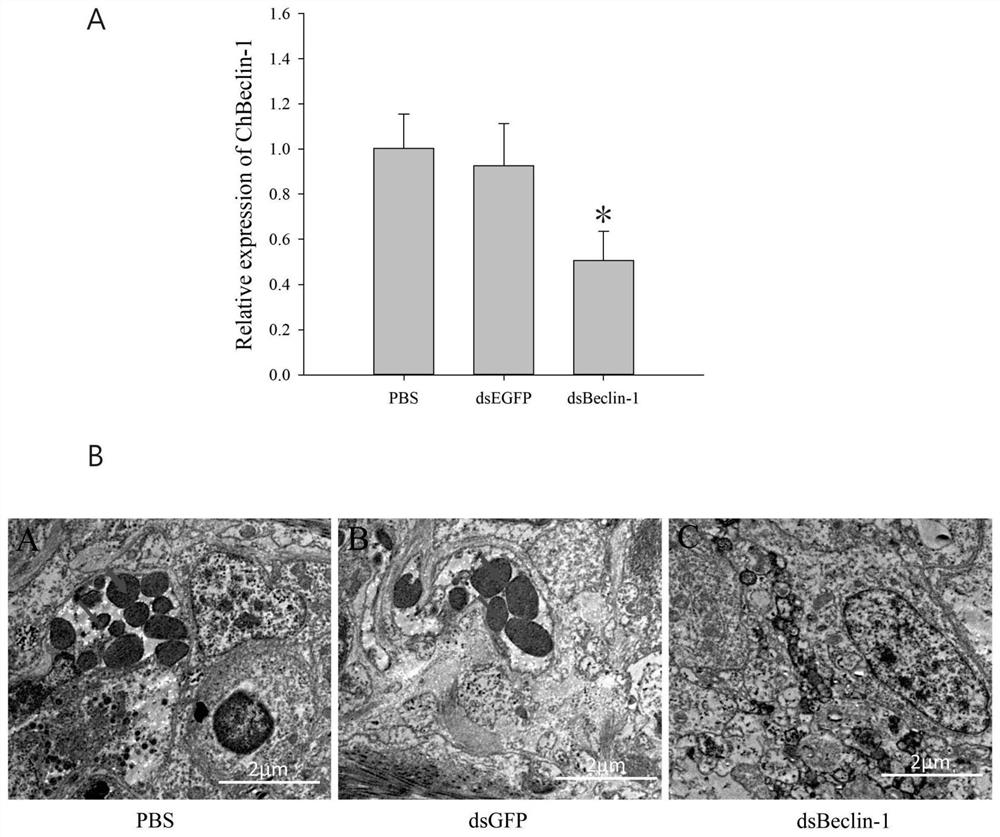
[0025] Example 3: Expression pattern of Hong Kong oyster Beclin-1 under bacterial infection
[0026] The expression pattern of Beclin-1 gene in Hong Kong oysters infected by bacteria was detected by real-time fluorescent quantitative method (qPCR). 200 Hong Kong oysters were equally divided into two groups (four groups), and Vibro alginolyticus (Vibro alginolyticus) and Staphylococcus aureus (Staphylococcus haemolyticus) were resuspended in PBS buffer so that the final concentration was 1×10 9 cfu / mL, with a 1mL syringe, inject 0.1mL of the above-mentioned concentration of Vibrio bacteria into the adductor muscle of Hong Kong oysters, and at the same time inject 0.1mL of PBS buffer into each oyster of the control group, and the blank group does not receive any treatment. 3, 6, 12, 24, 48 and 72 hours after the stimulation, 5 oysters were randomly selected from each group, the hemolymph was extracted, and the hemolymphocytes were collected by centrifugation at 800×g for 5 minut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com