Method for screening neoantigen or neoantigen coding sequence
A coding sequence, nascent technology, applied in biochemical equipment and methods, tumor-specific antigens, chemical instruments and methods, etc., can solve the problem of neoantigen peptide short peptides, cannot evaluate the degradation rate, and cannot achieve primary tumor cell killing verification and other issues to achieve the effect of good affinity and large proportion of mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] (1) Gene design and synthesis for screening neoantigens
[0041] First, we designed the genes for screening neoantigens, and added kozak, signal peptide and ubiquitin genes before the DNA sequence encoding neoantigens (neoantigen coding sequence). Among them, the kozak sequence can combine with the translation initiation factor to mediate the initiation of mRNA translation of the 5' cap structure, thereby increasing the gene expression. Since the assembly of peptides and MHC class I molecules occurs in the endoplasmic reticulum, in order to increase the collision probability between peptides and MHC class I molecules, we added signal peptide and ubiquitin genes before the neoantigen coding sequence to promote the amino acid sequence obtained by transcription and translation It can be cleaved by the proteasome near the endoplasmic reticulum, and the short peptides obtained after cleavage can be transported to the surface of the cell membrane by MHC class I molecules. Si...
Embodiment 2
[0073] (1) Gene design and synthesis of short gene tandem sequence (tandem minigene, TMG)
[0074] We inoculated 2 × 10 subcutaneously on the right side of the mouse 5 B16F10 cells were inoculated into the tail vein of mice with 1×10 5 B16F10 cells were obtained from mouse subcutaneous melanoma tissue and lung metastatic melanoma tissue. We sequenced the whole exome and transcriptome of subcutaneous melanoma, lung metastatic melanoma, and B16F10 cells. FastQC software filters the data with poor quality in the raw data, BWA software compares the sequencing data with the mouse reference gene mm10 downloaded from ensembl, and GATK software uses mutect1, strelka, varscan and sniper to analyze somatic mutations. In addition, we also analyzed mutations with copy number variation by CNVkit, FREEC and PyLOH software. Low-quality mutations in the mutation sites were removed, and the remaining mutations were used as somatic mutation sites for subsequent analysis. Screen the neoantig...
Embodiment 3
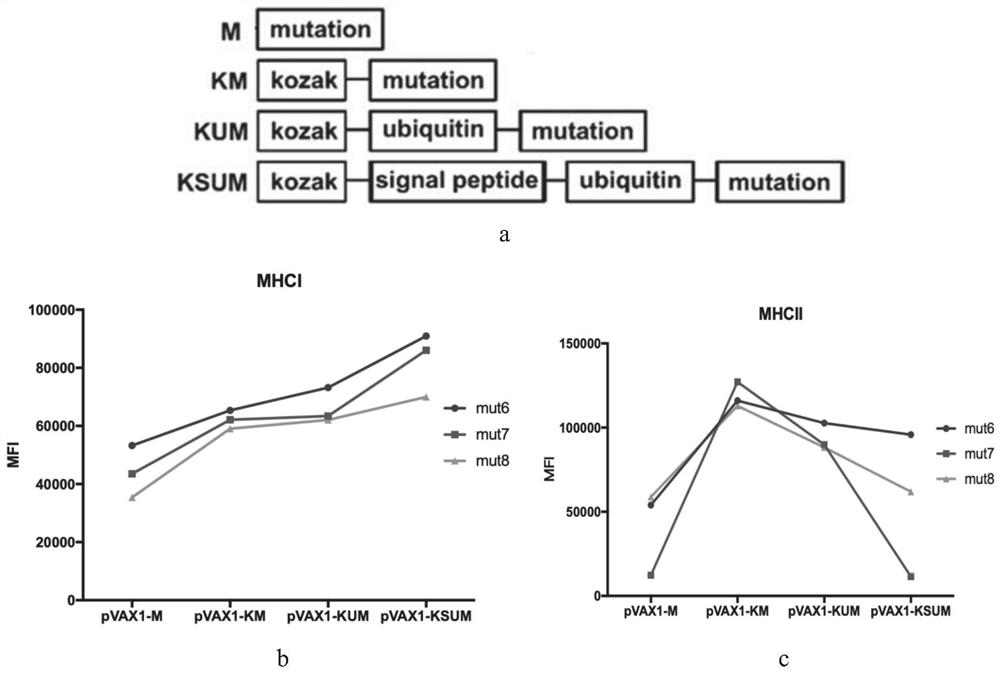
[0099] After determining the TMG sequence with strong immunogenicity, we further screened the neoantigens encoded on TMG one by one, and loaded the neoantigen coding gene, namely the mutation gene, on the pVAX1-KSU plasmid, in which the mutation gene encodes 25 amino acid long neoantigen, the mutation site is usually in the middle of the sequence.
[0100] (1) Gene construction for screening candidate neoantigens and corresponding wild-type antigen peptides
[0101] On the basis of the existing kozak-signal peptide-ubiquitin (KSU) gene, the neoantigen coding gene (mutation, M) or wild type antigen peptide (wild type, WT) gene was added to its 3' end by PCR. We loaded these different genetic elements on the pVAX1 plasmid for subsequent cell transfection.
[0102] (2) Screening of candidate tumor neoantigens
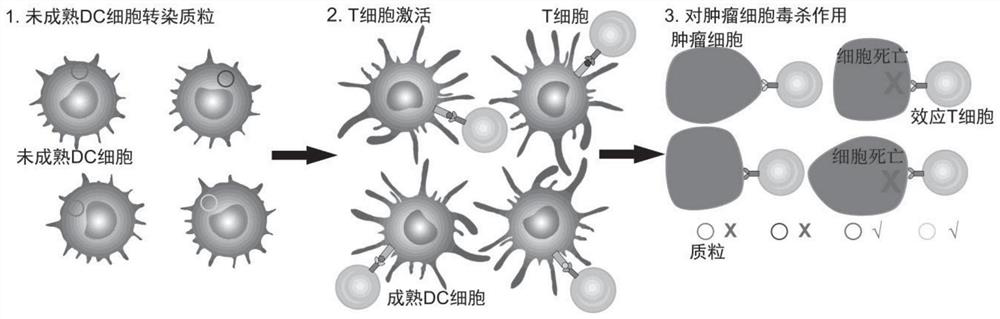
[0103] a. BMDC cells (mouse bone marrow-derived dendritic cells) were transfected with the pVAX1 plasmid loaded with neoantigen-encoding genes, centrifuged after 12 hour...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com