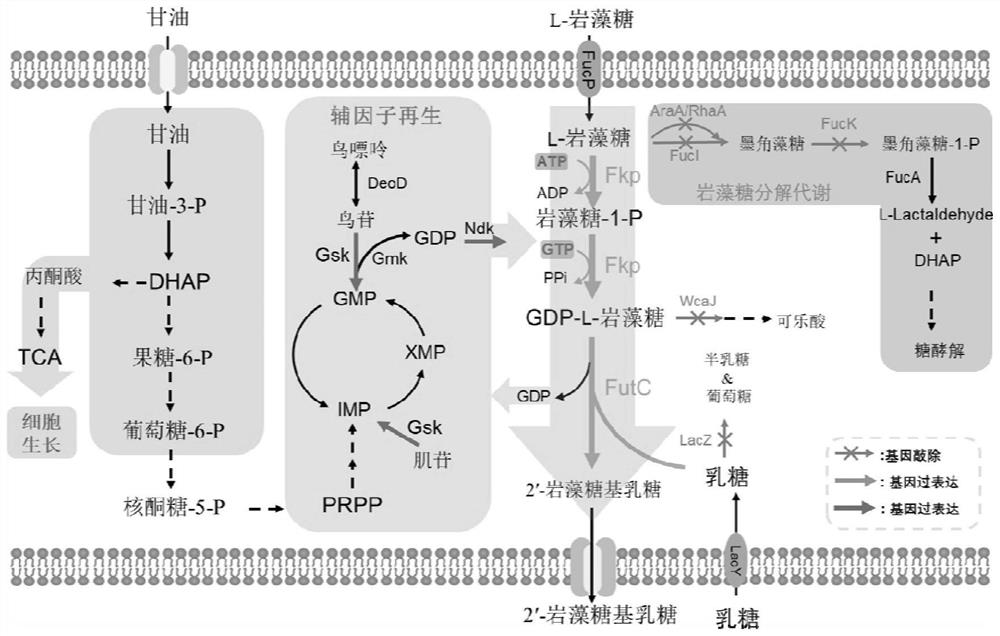
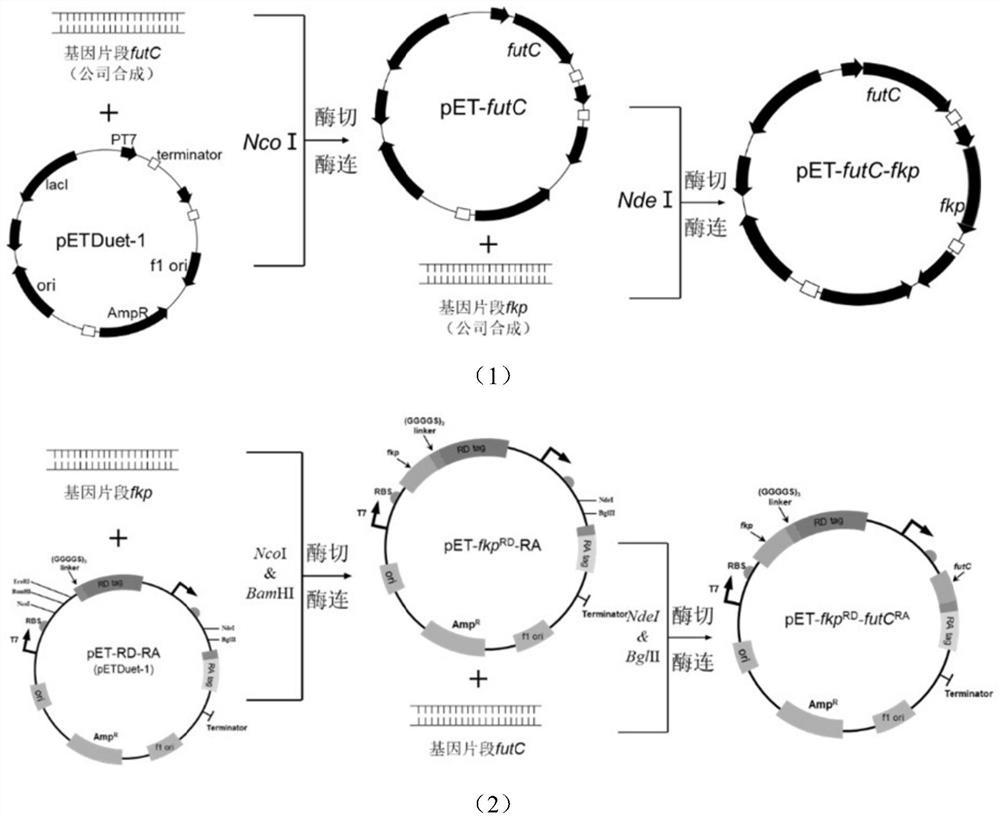
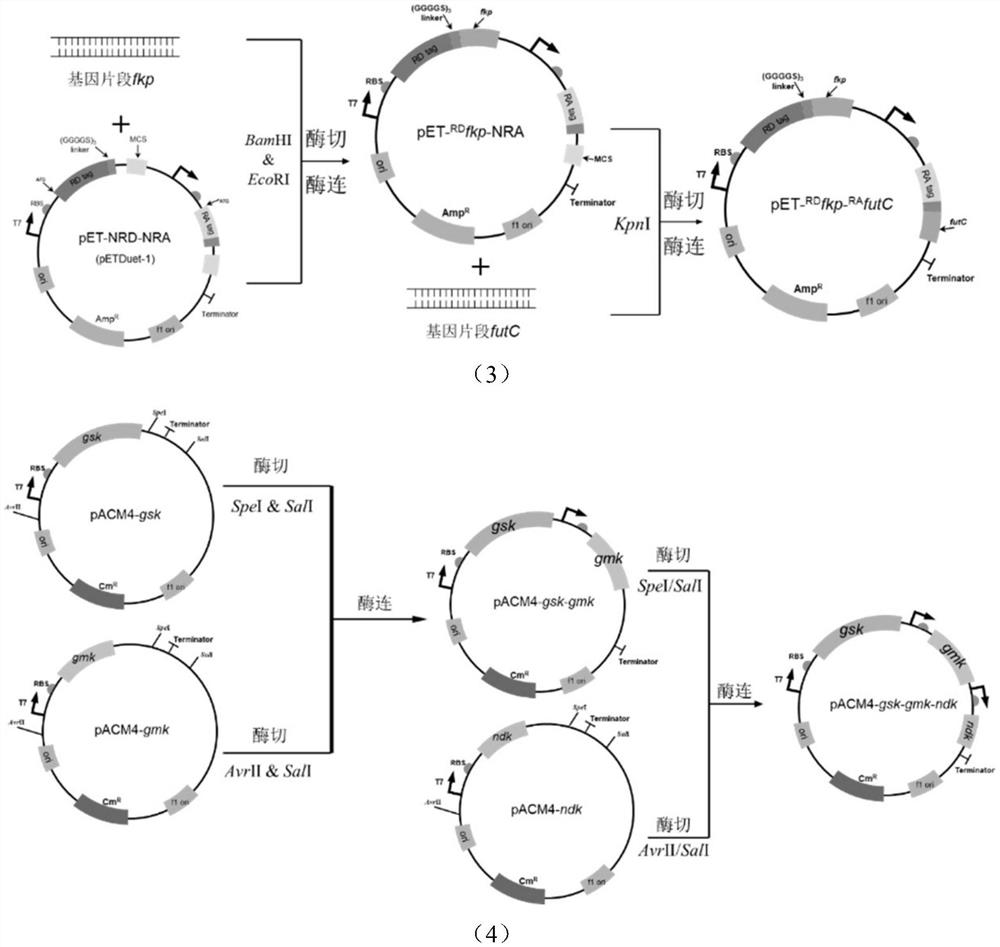
Escherichia coli engineering strain for efficiently producing 2'-fucosyllactose
A technology of fucosyllactose and Escherichia coli, applied in the fields of synthetic biology and microbial metabolic engineering, can solve the problems of short salvage pathway, low 2′-FL yield, and influence on host metabolism, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1: Construction of recombinant Escherichia coli knockout lacZ
[0079] (1) Use primer N20-ΔlacZ-F / R to perform PCR amplification on the existing pTargetF plasmid to obtain the pTargetF plasmid with the targeting lacZ gene;
[0080] (2) Use primers ΔlacZ-UH-F / R and ΔlacZ-DH-F / R to amplify upstream and downstream homologous fragments of the lacZ gene by PCR (see Table 6 for PCR primer sequences);
[0081] (2) These two homologous fragments pass The II one-step cloning kit was connected to the linearized pTargetF-ΔlacZN20 vector (the vector was linearized by primer pTargetT-F / R amplification) to construct the plasmid pTargetT-ΔlacZN20;
[0082] (3) Transfer the pCas plasmid into E.coliBL21(DE3) by electroporation, and add 20mM arabinose to induce the expression of the λ-Red Escherichia coli gene recombination system;
[0083] (4) Electroporate the above-mentioned Escherichia coli BL21(DE3) with the pTargetT-ΔlacZN20 plasmid, spread it on an LB plate containing ka...
Embodiment 2
[0086] Example 2: Construction of Escherichia coli expressing futC and fkp heterologously
[0087] The genes futC (nucleotide sequence shown in SEQ ID NO.2) and fkp (nucleotide sequence shown in SEQ ID NO.1) were synthesized by Shanghai Sangon Bioengineering Co., Ltd. The genes futC and fkp were respectively amplified by primers futC-F / R(NcoI) and fkp-F / R(NdeI) and then inserted into the NcoI restriction site of the first multiple cloning site of pETDuet-1 after restriction enzyme ligation point and the NdeI restriction site of the second multiple cloning site, constituting the plasmid pET-futC-fkp (see Table 1 for the primer sequence, and refer to the plasmid construction flow chart figure 2 (1)).
[0088] The plasmid pET-futC-fkp was transformed into E.coliBL21(DE3)ΔlacZ, and the strain containing plasmid pET-futC-fkp was obtained after sequencing verification, which was named WLS01.
Embodiment 3
[0089] Embodiment 3: the construction of the recombinant escherichia coli of knocking out fucIK, araA, rhaA, wcaJ gene
[0090] According to the method of Example 1, the fucIK gene in the bacterial strain WLS01 was knocked out, and the bacterial strain WLS02 was constructed;
[0091] According to the method of Example 1, the araA gene in the bacterial strain WLS02 was knocked out, and the bacterial strain WLS03 was constructed;
[0092] According to the method of Example 1, the rhaA gene in the bacterial strain WLS02 was knocked out, and the bacterial strain WLS04 was constructed;
[0093] According to the method of Example 1, the rhaA gene in the bacterial strain WLS03 was knocked out, and the bacterial strain WLS05 was constructed;
[0094] According to the method in Example 1, the wcaJ gene in the strain WLS05 was knocked out to construct the strain WLS06.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com