Glycoprotein N-sugar chain analysis method
An analysis method and glycoprotein technology, applied in the direction of material analysis, material analysis by electromagnetic means, material separation, etc., can solve problems such as the health threat of experimenters, unstable fluorescence signal, increase material cost, etc., to increase popularization, Strong applicability and the effect of increasing detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
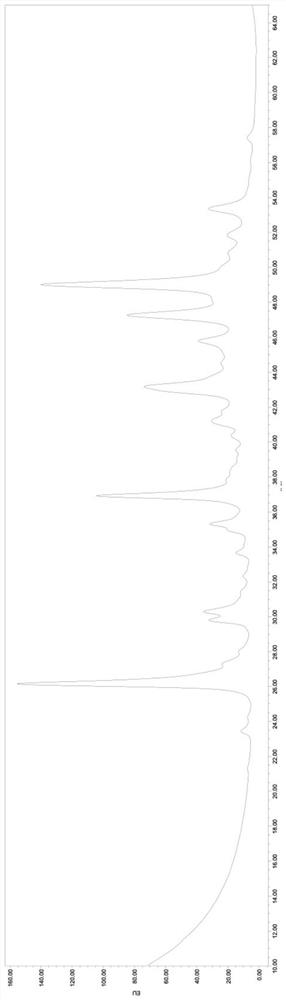
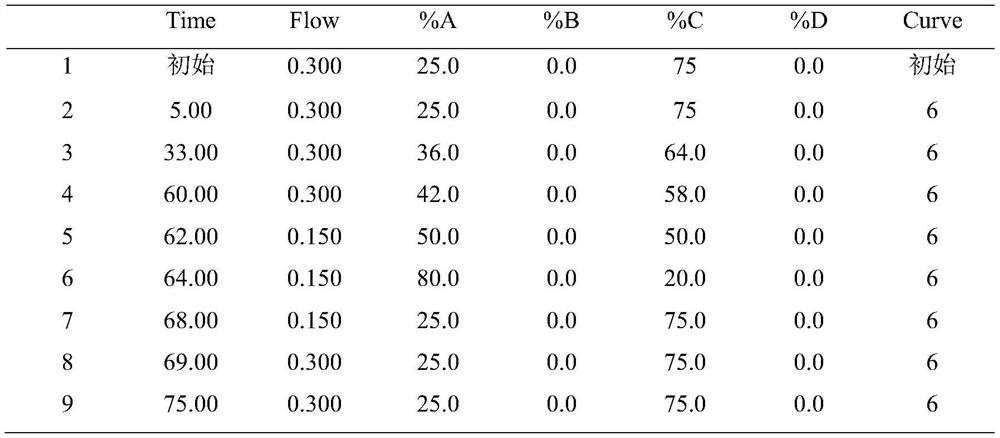
[0066] Take the recombinant human erythropoietin (Fc) fusion protein sample for inspection according to the following steps, and the obtained N-glycan spectrum chromatogram is shown in figure 1 .
[0067] 1) Sample dilution: Dilute the sample to 5 mg / ml with dilution buffer.
[0068] 2) Reduction: Take 15 μl of the diluted sample solution in a 1.5ml EP tube, add 15 μl of 2 mg / ml RapiGest SF solution, mix for 5 seconds, then add 1 μl of 0.5M DTT, mix well and put it in a 37°C incubator Restore for 30min.
[0069]3) Alkylation: after cooling to room temperature, add 2 μl of 0.5M IAM, mix well, and place in the dark at room temperature for 30 min.
[0070] 4) Enzyme digestion: add 4 μl PNGase F (250 U / ml), mix well, and place in a 37° C. incubator for enzyme digestion for 2 hours.
[0071] 5) Drying: Open the cover of the enzyme-digested sample and put it into a vacuum drying oven to slowly evacuate the sample to completely dry the sample.
[0072] 6) Labeling: 5 μl of the pr...
Embodiment 2
[0078] Human IgG samples were taken for testing according to the following steps.
[0079] 1) Sample dilution: Dilute the sample to 5 mg / ml with dilution buffer.
[0080] 2) Reduction: Take 15 μl of the diluted sample solution in a 1.5ml EP tube, add 15 μl of 3mg / ml RapiGest SF solution, mix for 5 seconds, then add 1 μl of 0.5M DTT, mix well and put it in a 34°C incubator Reduction 40min.
[0081] 3) Alkylation: after cooling to room temperature, add 2 μl of 0.5M IAM, mix well, and place in the dark at room temperature for 20 min.
[0082] 4) Enzyme digestion: Add 4 μl PNGase F (300 U / ml), mix well, and place in a 37°C incubator for enzyme digestion for 1.5 hours.
[0083] 5) Drying: Open the cover of the enzyme-digested sample and put it into a vacuum drying oven to slowly evacuate the sample to completely dry the sample.
[0084] 6) Labeling: 5 μl of the prepared labeling reagent was added to the aforementioned enzyme-digested sample, mixed evenly, and placed in a 65° C. ...
Embodiment 3
[0087] Serum transferrin samples were taken for testing according to the following steps.
[0088] 1) Sample dilution: Dilute the sample to 5 mg / ml with dilution buffer.
[0089] 2) Reduction: Take 15 μl of the diluted sample solution in a 1.5ml EP tube, add 15 μl of 1 mg / ml RapiGest SF solution, mix for 5 seconds, then add 1 μl of 0.5M DTT, mix well and put it in a 39°C incubator Restore for 20min.
[0090] 3) Alkylation: After cooling to room temperature, 2 μl of 0.5 M IAM was added, mixed well, and placed at room temperature in the dark for 40 minutes.
[0091] 4) Enzyme digestion: add 4 μl PNGase F (200U / ml), mix well, and place in a 37°C incubator for enzyme digestion for 2.5 hours.
[0092] 5) Drying: Open the cover of the enzyme-digested sample and put it into a vacuum drying oven to slowly evacuate the sample to completely dry the sample.
[0093] 6) Labeling: 5 μl of the prepared labeling reagent was added to the aforementioned enzyme-digested sample, mixed evenly,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com