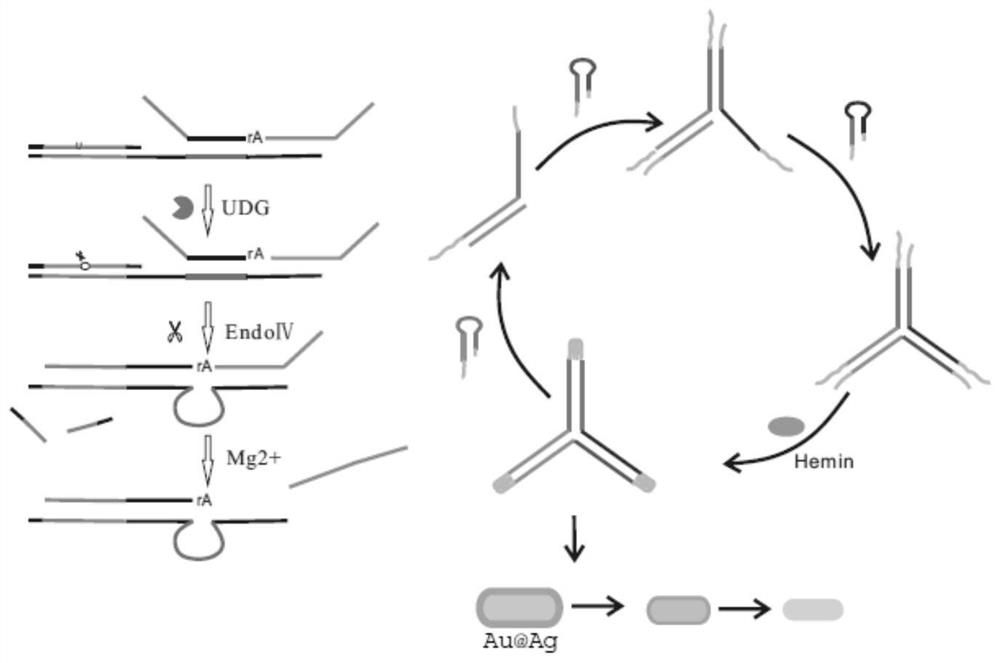
Colorimetric biosensor for detecting UDG based on Au@Ag
A biosensor and colorimetric technology, applied in the field of biosensors, can solve the problems of genome stability and accuracy interference, harmful cells, carcinogenicity, etc., and achieve the effects of fast detection speed, pollution avoidance, and low detection limit
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Preparation of Au@Ag
[0045] 1. Preparation method of gold rod
[0046] 1.1, the preparation method of gold seed
[0047] Mix 5 mL of CTAB solution (0.2 M) with 5 mL of HAuCl4 solution (0.0005 M) and invert several times. Freshly prepared NaBH 4 (0.01M) 0.6 mL was added to the above mixed solution, shake vigorously for 2 min, and store in a water bath at 25°C for 1 h.
[0048] 1.2. Preparation method of growth solution
[0049] Mix 100 mL CTAB solution (0.2 M) with 6 mL AgNO 3 (0.0035 M) mixed, HAuCl 4 (0.01M) 100 mL was added to the above mixed solution and mixed gently. Add 1.4 mL (0.0788M) of ascorbic acid, and the color of the solution changes from dark yellow to colorless rapidly. Finally, add 240 μL of seed solution to the growth medium at 28 °C. Take it out after 8 hours and centrifuge at 8500 rpm for 10 minutes.
[0050] 1.3. Preparation method of Au@Ag
[0051] The AuNRs solution was centrifuged at 8500 rpm for 10 min, and then resuspend...
Embodiment 2
[0052] Example 2 S 123 Triplex preparation
[0053] Synthesize the required DNA triplex according to the sequence shown in No:1-3, and pass through Tris buffer (50 mm Tris, 100mm NaCl, 50 mm KCl, 1 mm MgCl 2 , pH 7.4) to prepare S by heating and annealing S1 / S2 / S3 123 , the solution was heated at 90 °C for 5 min and then slowly cooled to room temperature.
Embodiment 3
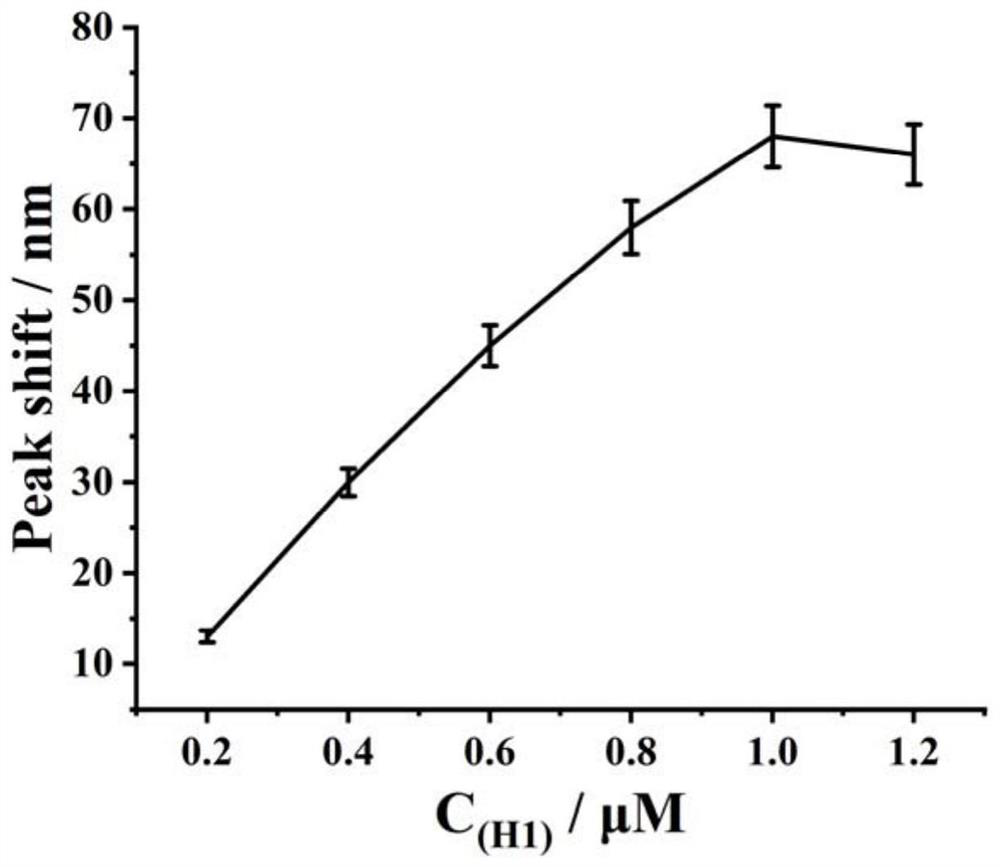
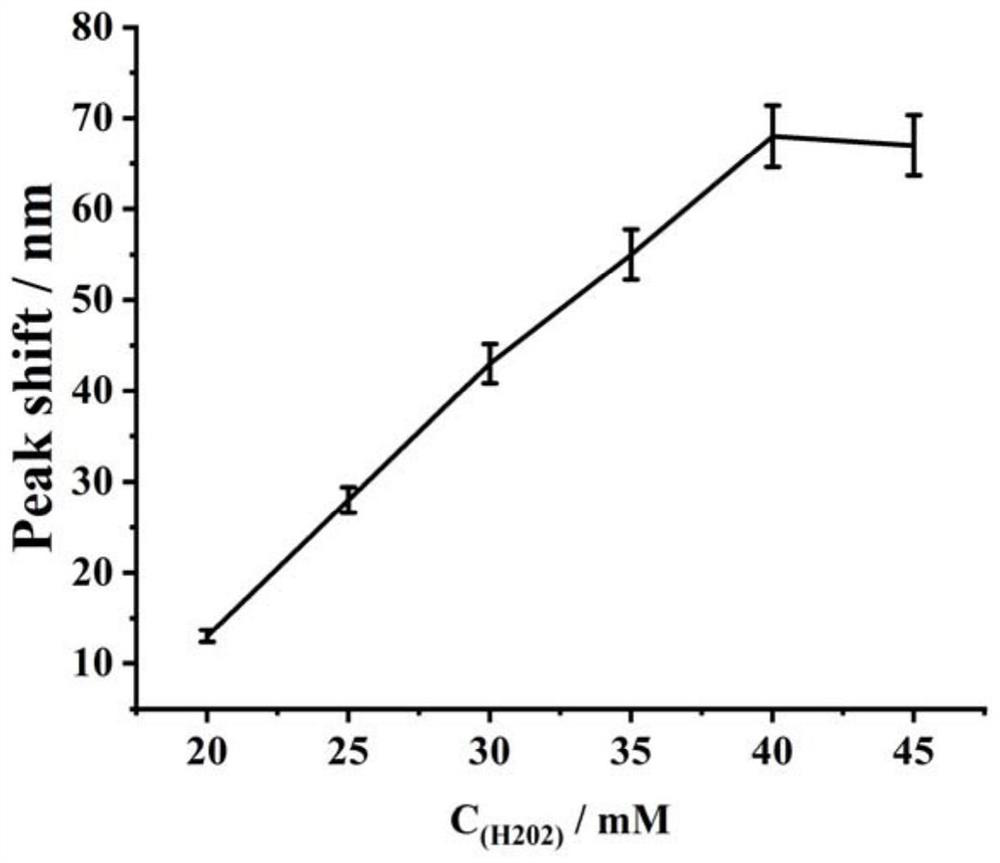
[0054] Example 3 Effect of H1 concentration change on exosome detection
[0055] Take the triplex synthesized in Example 2, hairpin probes H1, H2, and H3, and then screen the optimal concentration of H1 according to the following steps:
[0056] (1) Take 6 EP tubes and add 10 μL S 123 (1 μL, 5 μM), UDG (1 μL, 1U / μL), 2 UEndoⅣ, 50 mM Tris-HCl buffer (100 mm NaCl, 50 mM KCl, 1 mM MgCl 2 , pH 7.4 (pH 7.4)) mix, 37°C, react for 2 h;
[0057] (2) Then add equal volumes of H1 solutions with different concentrations, so that the final concentrations are 0.2 μM, 0.4 μM, 0.6 μM, 0.8 μM, 1 μM, 1.2 μM, and then add H2 (1 μL, 1 μM), H3 (1 μL, 1 μM), 37°C, react for 90min;
[0058] (3) Then add heme (1 μL, 1 μM) and react at 37°C for 30 min; then add H 2 o 2 (2 μL, 40 mM), and finally add 15 μL Au@Ag (Abs. 0.5) for detection by UV-Vis scanning absorbance, the scanning range is 400-800 nm.
[0059] The result is as figure 2 As shown, it can be seen from the figure that the detected ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com