Method for rapidly and quantitatively detecting enterobacter sakazakii based on cascade amplification
A technology for quantitative detection of Enterobacter sakazakii, applied in biochemical equipment and methods, measurement/inspection of microorganisms, resistance to vector-borne diseases, etc., to achieve the effect of ensuring specificity, high biological activity, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
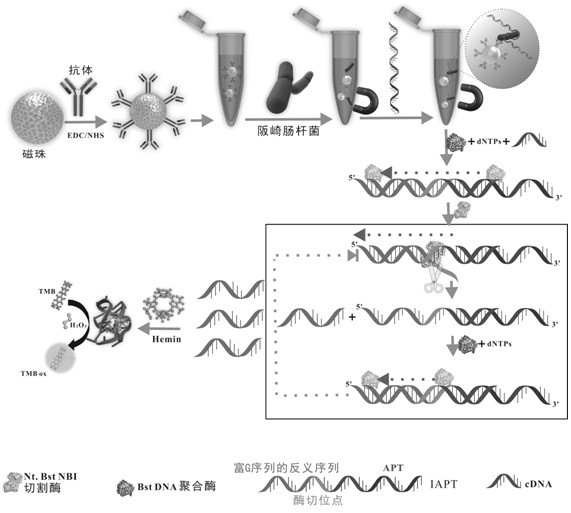
[0046] Example 1 Principle of rapid detection method for Enterobacter sakazakii
[0047] The principle of rapid quantitative and visual detection of Enterobacter sakazakii figure 1 As shown, first, the classic EDC / NHS method is used to construct immunomagnetic beads, that is, to couple antibodies to the magnetic beads; when the target exists, the antibody can be used as a capture recognition element, and then the fusion nucleic acid aptamer (IAPT) is added to form Highly specific sandwich structure. In addition, since the fusion nucleic acid aptamer contains the amplification template of EXPAR, Nt.BstNBI Dicer recognition site and G-rich antisense sequence, so in Bst. Under the action of DNA polymerase, EXPAR can be triggered, and the product contains a large number of G-rich sequences; this sequence combines with Hemin to form G4 / HeminDNAzyme, which catalyzes H 2 o 2 The colorless TMB is oxidized to blue, and finally the experimental results can be evaluated by the di...
Embodiment 2
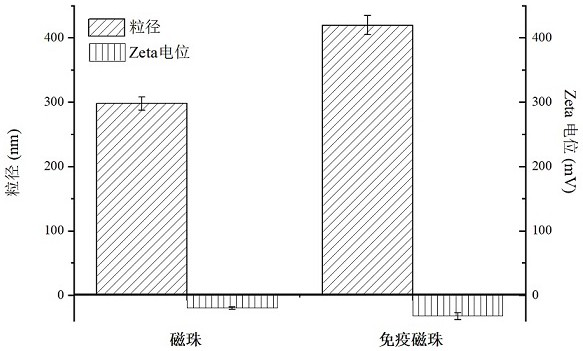
[0051] Example 2 Construction of immunomagnetic beads and related characterization
[0052] Carboxyl magnetic beads were washed with 500 μL of reaction buffer (0.05 M 2(N-morpholine)ethanesulfonic acid, 0.5 M NaCl, pH=6.0) for three times and then magnetically separated and activated by classical EDC / NHS method. Then add 200 μL of coupling solution (0.1M sodium phosphate, 0.15 M NaCl, pH=7.5) to wash the activated magnetic beads three times, and remove the supernatant after separation. Dissolve 0.5 mg antibody in 0.5 mL coupling solution as stock solution, add antibody to magnetic beads according to the ratio of magnetic beads:antibody = 10:1, shake and couple at room temperature for 2 h, perform magnetic separation to remove supernatant, and then add 200 μL of blocking solution (add 1 M glycine to the coupling solution, pH=8.5), shake at room temperature for 30 min, block unreacted activating groups, and finally dilute to working concentration with preservation solution and s...
Embodiment 3
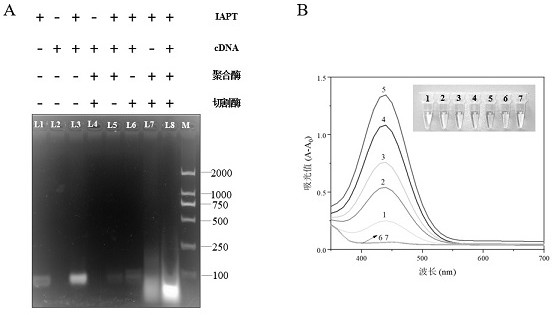
[0054] Example 3 Feasibility of detection of Enterobacter sakazakii
[0055] After the immune sandwich structure was constructed, the EXPAR reaction was carried out, and the specific EXPAR reaction system is shown in Table 2. In order to verify the feasibility of using cascade amplification to detect Enterobacter sakazakii, the occurrence of EXPAR amplification was first verified by gel electrophoresis.
[0056] Table 2 EXPAR reaction system
[0057]
[0058] The result is as image 3 As shown in A, the clear band in lane 1 represents IAPT while the cDNA (lane 2) has no obvious band due to its short single strand. Lane 3 represents the hybridization result of IAPT and cDNA, compared with lane 1, its mobility is reduced, indicating that the two hybridize successfully. Only when IAPT, cDNA, Bst . DNA polymerase, Nt.BstNBI The characteristic band of lane 8 will appear only when cleavage enzyme and dNTP co-exist, which indicates the generation of short DNA fragments (i.e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com