Protein glutaminase production strain, screening method and characteristic analysis method
A glutaminase and protein technology, applied in the field of bioengineering, can solve the problems of low enzyme production capacity, protein aggregation, easy to produce bitter taste, etc., and achieve the effects of high enzyme production and rapid growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 Screening of protein-producing glutaminase strains
[0066] (1) Soil suspension enrichment culture
[0067] The screening steps are:
[0068] Add 90mL of distilled water and 20-30 glass beads into a 250mL shaker flask, and sterilize by high pressure steam at 121°C for 20min. In an ultra-clean workbench, weigh 10 g of soil sample, add it to a shaker flask filled with distilled water, shake at 30° C. and 200 rpm / min for 30 min, and prepare a 10% (w / v) soil suspension. Then place it in an ultra-clean workbench and let it stand. Take 1% (v / v) soil suspension into the enrichment medium (60 mL), shake at 30° C. and 200 rpm / min for 5 days.
[0069] The enrichment medium is: lactose 5g, yeast powder 11.7g, CBZ 1g, K 2 HPO 4 0.1g, KH 2 PO 4 0.1g, 0.1g sodium chloride, pH7.2, add distilled water to make up to 1L, and sterilize at 115°C for 20min.
[0070] (2) Plate primary screening
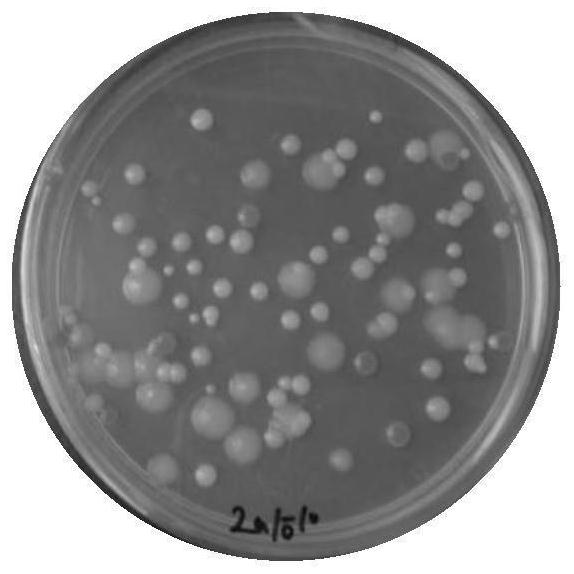
[0071] Dilute the enriched bacterial solution to multiple ratios, select an...
Embodiment 2
[0085] The identification of embodiment 2 protein-producing glutaminase strains
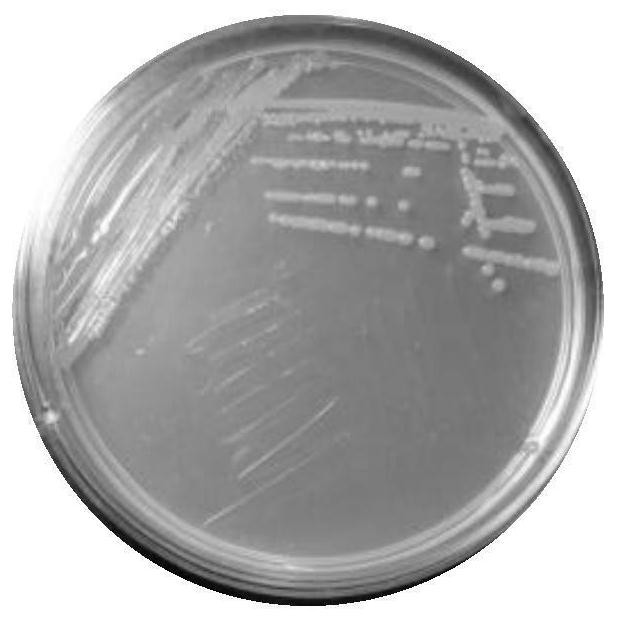
[0086] The suspected strain screened in Example 1 of the present invention was re-streaked on the LB plate to obtain a single colony, and then the color and state of the strain were observed. The colony was round, the colony was full, the edges were smooth, no nicks, and the colony was sticky. Such as image 3 shown.
[0087] Subsequent Gram staining revealed that the strain was rod-shaped, non-spore-forming, and Gram-negative.
[0088] According to the instructions of the physiological and biochemical identification tube, the first choice is to activate the strain, then prepare a bacterial suspension, inoculate it in the physiological and biochemical tube and cultivate it for 24 hours, observe the experimental phenomenon and make a record. Among them, "+" indicates that the strain is positive, and "-" indicates that the strain is negative. The identification results were different from the Ch...
Embodiment 3
[0095] Identification and Analysis of Protein Glutaminase of Example 3 Bacterial Strain A4142
[0096] Extract the total genomic DNA of bacterial strain A4142, and use this as a template to amplify the full-length DNA sequence of the protein glutaminase produced by the bacterial strain, and its primer sequence is:
[0097] SEQ ID No. 3: F-proteolyticum AACTTGCTTATGTTATTTTTTTTTAT
[0098] SEQ ID No.4: R-proteolyticum GGATGTTATCATACAAAAAAAATAAT
[0099] The PCR products were detected by 1% agarose gel electrophoresis, recovered by tapping the gel, and sent to Shanghai Sunny Company for sequencing. Obtain the gene sequence of the PG enzyme of bacterial strain A4142, i.e. SEQ ID No.5 (963bp in total in the DNA sequence), translate the sequence to obtain the PG enzyme protein sequence, shown in SEQ ID No.6, wherein there are 320 protein sequences in total Amino acids, wherein 21 amino acid signal peptides (as shown in SEQ ID No.7), 114 amino acid propeptides (as shown in SEQ ID N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com