Dual fluorescent quantitative PCR (polymerase chain reaction) detection method and kit for two pathogenic bacteria in prepackaged drinking water
A technology for fluorescence quantification and detection reagents, which is applied in the field of fluorescence quantitative PCR detection, can solve the problems of long detection time and the detection limit cannot meet the limit requirements of pathogenic bacteria, and achieves low reagent cost, high practical value and good stability. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
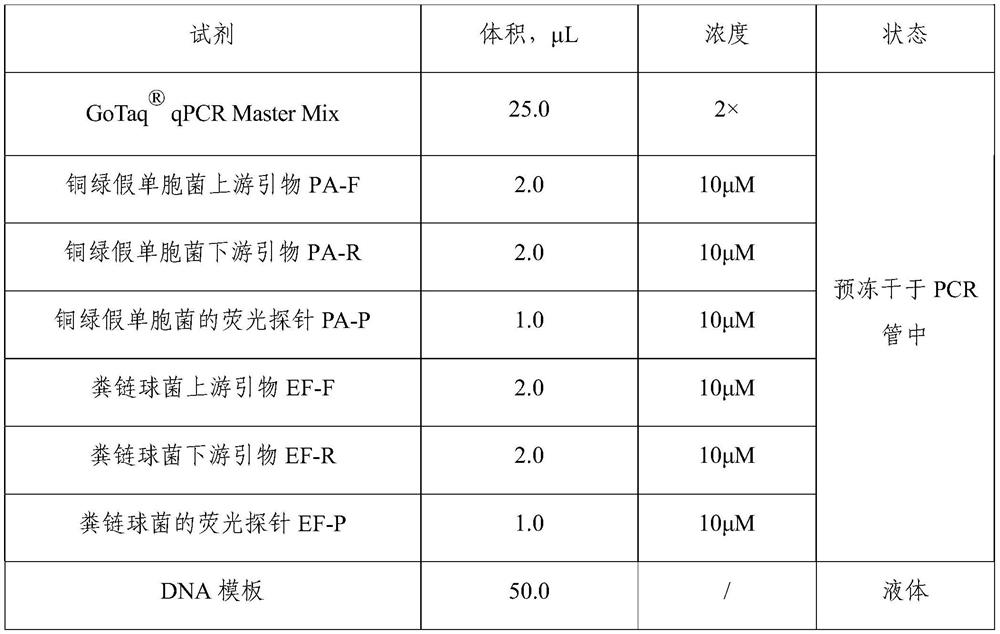
[0063] Embodiment 1 detects the construction and verification of the double fluorescent PCR kit of two kinds of pathogenic bacteria (pseudomonas aeruginosa and faecal streptococcus) in drinking water
[0064] 1. Design of primers and probes
[0065] According to the Pseudomonas aeruginosa-specific gyrB gene coding region sequence published by GenBank, specific primers and probes for Pseudomonas aeruginosa were designed, and the sequences are as follows (SEQ ID NO:1-3):
[0066] Upstream primer PA-F: 5'-GGCGTGGGTGTGGAAGTC-3'
[0067] Downstream primer PA-R: 5'-AGAACCTGCTCTGCTTCACCA-3'
[0068] Fluorescent probe PA-P: 5'-FAM-TGCAGTGGAACGACA-MGB-3'
[0069] According to the sequence of the 23S rDNA gene coding region of Streptococcus faecalis retrieved from GenBank, specific primers and probes for Streptococcus faecalis were designed. The sequences are as follows (SEQ ID NO: 4-6):
[0070] Upstream primer EF-F: 5'-GAGGACCGAACCCACGTA-3'
[0071] Downstream primer EF-R: 5'-CAGT...
Embodiment 2
[0081] Embodiment 2 Specificity and Sensitivity Investigation
[0082] 1. Specificity test
[0083] Using the primer set and probe set of Example 1, Pseudomonas aeruginosa and Streptococcus faecalis DNA are used as positive controls, and Salmonella, Bacillus cereus, Listeria, Proteus, Micrococcus luteus, Staphylococcus aureus The DNA of cocci, Clostridium perfringens, Citrobacter, and Escherichia coli were used as negative controls, and were detected with PCR kits to evaluate the specificity of the established fluorescent PCR method.
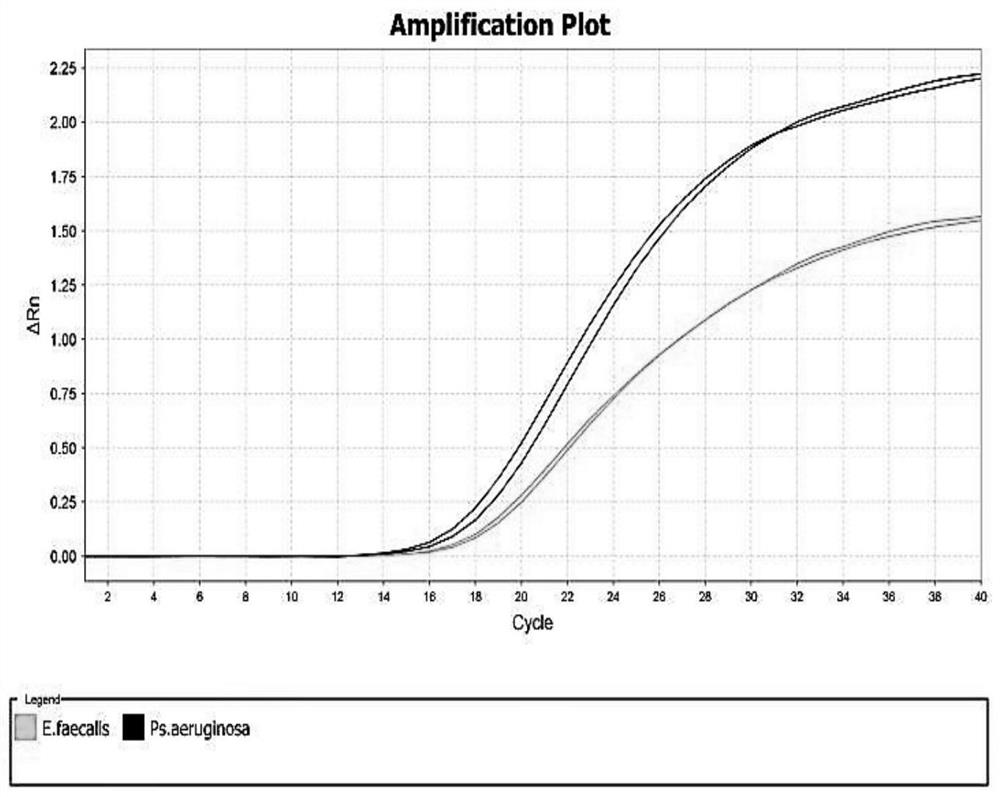
[0084] Experimental results such as figure 1 As shown, except Pseudomonas aeruginosa and Streptococcus faecalis with amplification curves and Ct value <30, other animal-derived templates had no amplification. It can be seen that the primers and probes provided by the present invention have good specificity and no cross-reaction with other microbial species.
[0085] 2. Sensitivity test
[0086] ①Pick the colonies of Pseudomonas aeruginosa an...
Embodiment 3
[0090] Example 3 Evaluation of the DNA Precipitation Effect of the DNA Precipitation Agent
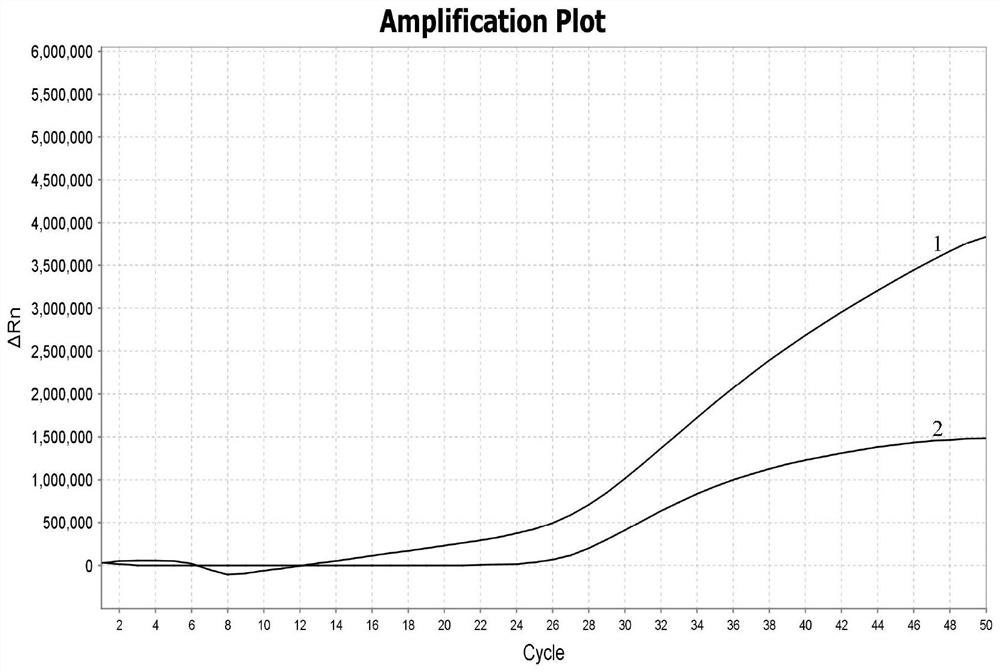
[0091] Taking Pseudomonas aeruginosa as an example, take 10 4 Dilute bacterial suspension 1.0mL, add to 250mL sterilized water. After filtration, the bacterial DNA on the filter membrane was extracted. Two filter membranes were used to extract DNA and detected according to the aforementioned method, and the other two filter membranes were used to extract DNA and detected according to the aforementioned method (without adding RNA carrier). The result is as figure 2 shown. The Ct value of the sample with RNA carrier filter membrane was 16.6, and the Ct value of the sample without RNA carrier filter membrane was 27.1. Therefore, the yield of cell DNA can be improved by adding RNA carrier.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com