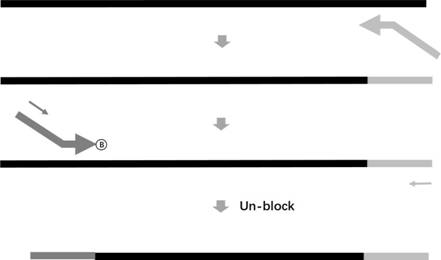
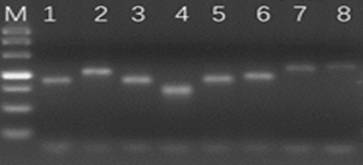
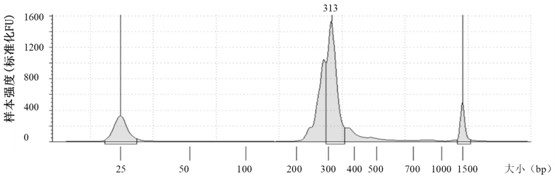
Method for determining minimal residual lesion level based on multiple amplification sequencing
A micro-residue, multiple amplification technology, applied in biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc., can solve the problem of ineffective removal of PCR preference, inaccurate and effective MRD level detection Problems such as low data ratio, to avoid the generation of primer-dimers, reduce UMI overlap, and improve efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0093] Example 1: Plasmid DNA Primer Amplification Test
[0094] Design and verification of target binding primers: download the reference sequences of immunoglobulin IgH, IgK, and IgL from public databases, and screen the homologous conserved regions through sequence comparison analysis for primer design. A total of 25 specific primers were designed. SEQ ID NO .1~25:
[0095] SEQ ID NO. 1 (IgHV1): ACAGYCTACATGGAGCTGAG
[0096] SEQ ID NO. 2 (IgHV2): GACCAACATGGACCCTGTGGA
[0097] SEQ ID NO. 3 (IgHV3): GGGCCGRTTCACCATCTCCA
[0098] SEQ ID NO. 4 (IgHV4): ACAGCCTGAAAACCGAGGACA
[0099] SEQ ID NO. 5 (IgHV5): CATCTCCAGAGACAATTCCARGAAC
[0100] SEQ ID NO. 6 (IgHV6): GCTGAACTCTGTGACTCCCGAGG
[0101] SEQ ID NO. 7 (IgHV7): GACACCTCTGCCAGCACAGCAT
[0102] SEQ ID NO. 8 (IgHJ): CTCACCTGAGGAGACAGTGACC
[0103] SEQ ID NO.9 (IgKV1): GGTTCAGYGGCAGTGGATCT
[0104] SEQ ID NO. 10 (IgKV2): ACWGATTTYACACTGAAAATCAGC
[0105] SEQ ID NO. 11 (IgKV3): CAGGCTCCTCATCTATGRTGCATC
[0106] SEQ ID NO...
Embodiment 2
[0129] Example 2: Cell Line DNA Studies
[0130] In this embodiment, the verification of MRD level detection is firstly performed on cell lines known in the public database. Specific steps are as follows:
[0131] 1. Screen cell lines:
[0132]
[0133] 2. Cell line nucleic acid preparation: All 6 cell lines were purchased from ATCC. Take 1mL of cell suspension and use the whole blood DNA extraction kit of Meijie to extract nucleic acid according to the instructions. The extracted nucleic acid was concentrated using Qubit3.0 and Agilent2100 and purity determination, and stored at -20°C for future use.
[0134] 3. Make the primer pool: refer to the primer sequence of SEQ ID NO.26~50, mix the 25 synthesized primers according to a certain ratio to obtain the forward primer pool and the reverse primer pool, of which SEQ ID NO.26~46 It is a forward primer (with a blocking modification group C3 at the 3' end), and SEQ ID NO.47~50 is a reverse primer. The number of forward prim...
Embodiment 3
[0212] Embodiment 3: clinical sample research
[0213] It has been verified in Example 2 that the detection result of the method for measuring MRD level of the present invention is accurate and effective for known cell lines, and this example uses clinical samples as materials for further verification. Specific steps are as follows:
[0214] 1. Clinical sample collection:
[0215] Samples of 15 patients with hematological malignancies were collected from cooperative hospitals. They may suffer from different diseases. The sample types include bone marrow fluid and peripheral blood. The patients were between 3 and 74 years old. There were 8 males and 7 females, see Table 2.
[0216] Table 2
[0217]
[0218] 2. Sample processing:
[0219]All 15 samples (bone marrow fluid and peripheral blood) were extracted using the Maijie Whole Blood DNA Extraction Kit according to the operating instructions.
[0220] 3. Amplified sequencing analysis: the steps are the same as in Exampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com