Solid phase nucleic acid detection probe and preparing method thereof
A technology for detecting probes and oligonucleotide probes, which is used in biochemical equipment and methods, microbial determination/inspection, fluorescence/phosphorescence, etc. The problems of mutation detection and single base mismatch detection are difficult to achieve the effect of improving the mismatch recognition ability, shortening the diagnosis time, and reducing the detection cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
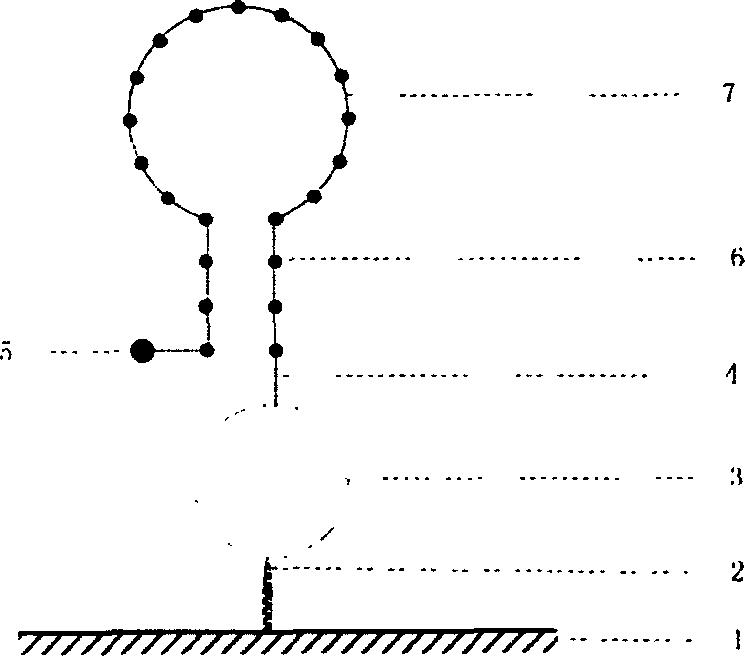
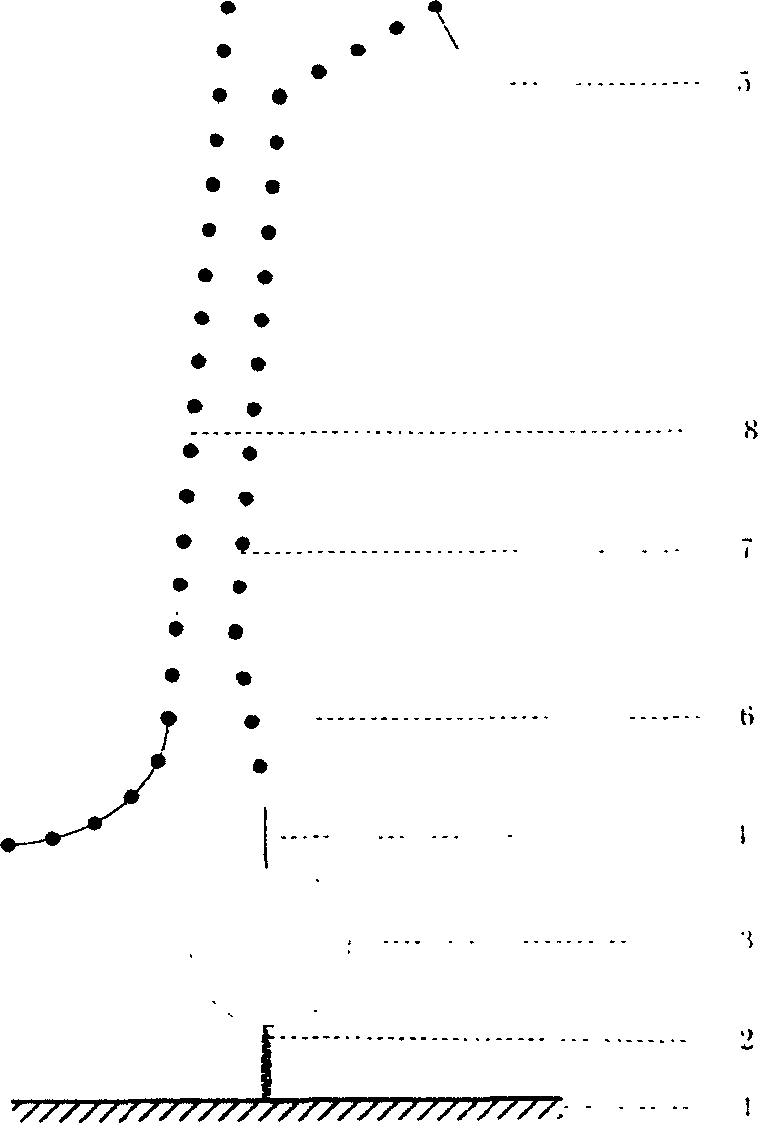
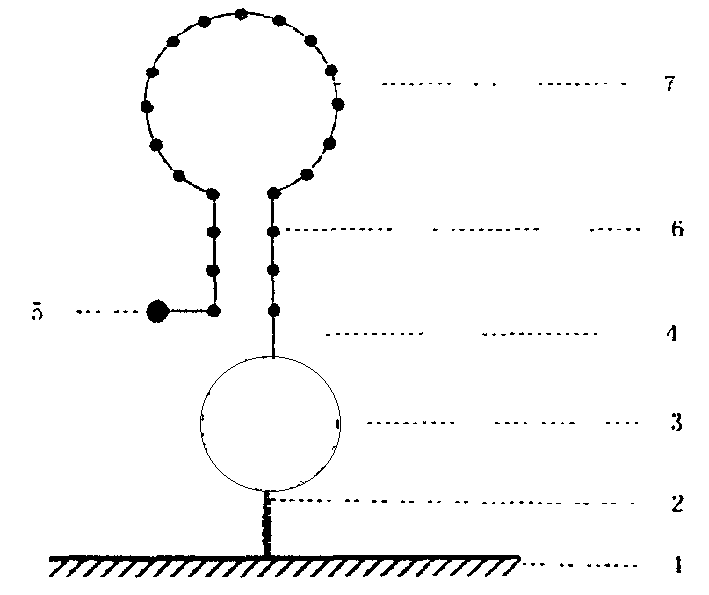
[0022] 4. Preparation of solid-phase nucleic acid probes: a commercial solid-phase chemical synthesis method was used to synthesize the equipment
[0023] A well-planned oligonucleotide probe comprising at least one fluorescent chromophore-specific nucleic acid sequence; the method for preparing the nucleic acid probe can also be to directly synthesize nucleic acid on a fluorescence quenching material by using an in-situ synthesis method and fluorescent labeling.
[0024] 5. Immobilization of probes: The probes synthesized by solid-phase chemistry are transferred to the surface of the solid substrate by machines or other means, and connected to the solid substrate under appropriate conditions. Each dot contains at least two probes labeled with different fluorophores.
[0025] 6. Hybridization and detection: Appropriate ions and buffers are added to the tested system, and the target gene and the solid-phase probe of the present invention are subjected to hybridization reaction, ...
Embodiment 1
[0026] Example 1, in situ synthetic non-marker gene chip
[0027] 1. Cleaning of glass slides: Soak the slides in lotion overnight, rinse, then soak in alkali ethanol solution for two hours, rinse with double distilled water, and blow dry with nitrogen for later use.
[0028] 2. Glass slide modification: Take clean slides, soak them in the acetone solution of triethoxyaminosilane for 5 minutes, wash them, bake them at 100 degrees for 40 minutes, soak them in glutaraldehyde solution for 2 hours, and wash them with nitrogen. blow dry.
[0029] 3. Gold nanoparticles immobilization: Soak the glass slide overnight in the gold nanometer solution modified with mercaptoethylamine, wash and blow dry with nitrogen.
[0030] 4. Synthesis of oligonucleotide probes: use the above-mentioned glass slides to synthesize nucleic acid sequences in an anaerobic and water-free glove box, and use the molecular stamp method to synthesize various sequences to make a chip, in which the last base is f...
Embodiment 2
[0031] Example 2, the immobilization of synthesized oligonucleotide probes and the production of non-labeled gene chips
[0032] 1. Cleaning of glass slides: Soak the slides in lotion overnight, rinse, then soak in alkali ethanol solution for two hours, rinse with double distilled water, and blow dry with nitrogen for later use.
[0033] 2. Glass slide modification: Take clean slides, soak them in the acetone solution of triethoxyaminosilane for 5 minutes, wash them, bake them at 100 degrees for 40 minutes, soak them in glutaraldehyde solution for 2 hours, and wash them with nitrogen. blow dry.
[0034] 3. Nano-gold immobilization: Soak the glass slide overnight with mercaptoethylamine-modified nano-gold solution, wash and blow dry with nitrogen.
[0035] 4. Synthesis of oligonucleotide probes: synthesized by conventional methods, one end of the oligonucleotide probe was modified with amino group, and the other end was modified with fluorescein.
[0036] 5. Chip fabrication:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com