Sodium alginate catenase gene algl and its preparation method and application
A alginate lyase and gene technology, applied in the direction of lyase, botany equipment and methods, biochemical equipment and methods, etc., can solve the problems that hinder the large-scale preparation of alginate oligosaccharides and hinder the application and development of the structure-activity relationship In-depth progress, unable to realize industrial production and other problems, to achieve the effect of good temperature and pH stability, low cost, and easy purification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
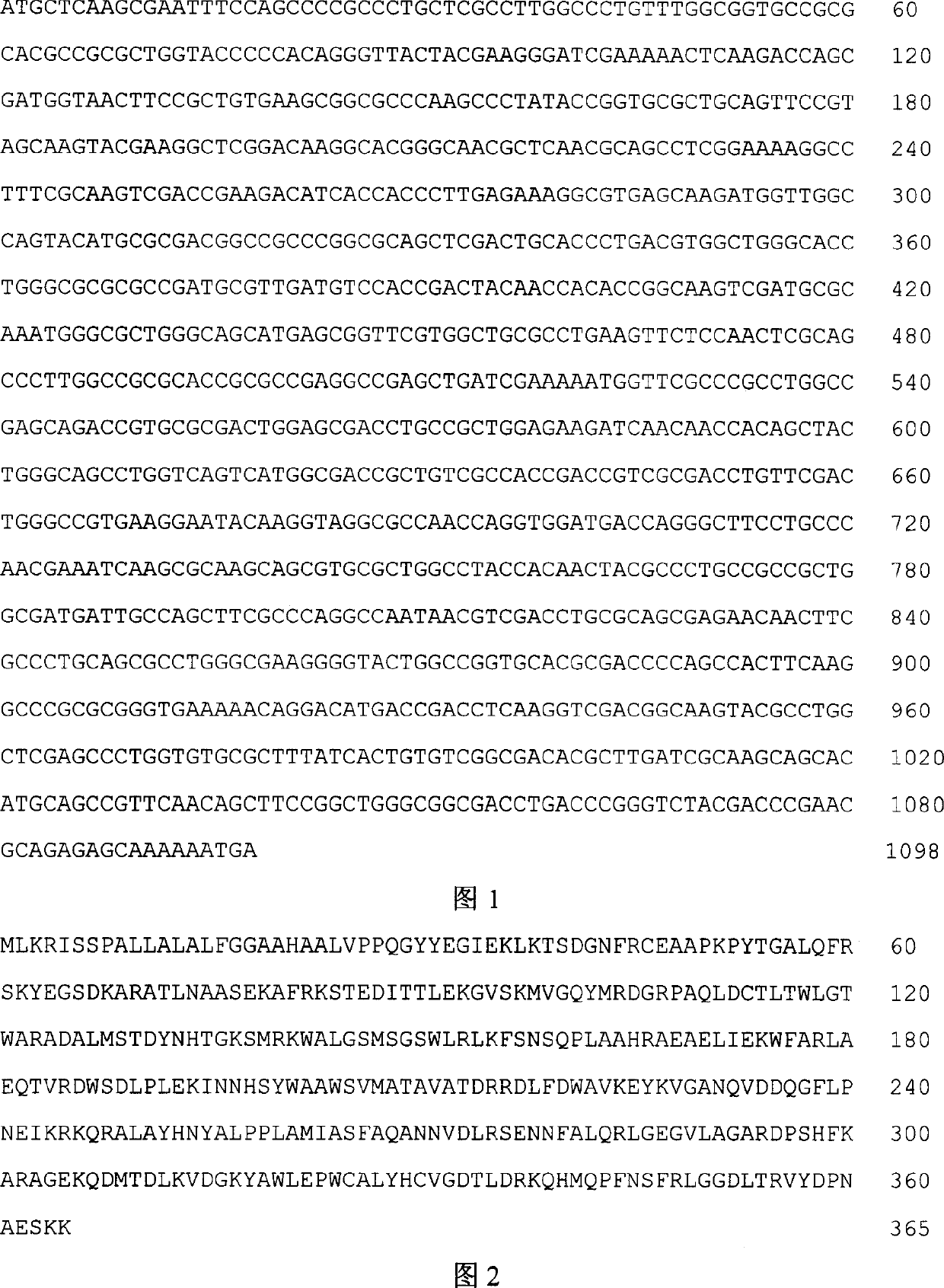
[0016] Example 1 Cloning of alginate lyase gene algL
[0017] Use degenerate polymerase chain reaction (PCR) primers to screen the genome library of Pseudomonas sp.) QDA, and clone the alginate lyase gene algL. The specific operation is: in Luria-Bertani ( Pseudomonassp.QDA was cultured in LB) medium to the end logarithmic phase, and genomic DNA was extracted. The genomic DNA was digested with Sau3AI, electrophoresed on agarose gel, and a 5-8 kb fragment was recovered, which was ligated with the pBluescript II KS (+) plasmid fragment that was completely digested with BamHI and dephosphorylated. Then, the ligation product was transferred into Escherichia coli DH5α competent cells according to the standard calcium chloride method, and cultured on LB solid medium (containing ampicillin, X-gal, IPTG). Pick the white spot recombinant colony, culture it in LB liquid medium (containing ampicillin), and use the degenerate primer pair UpperA (5'AACCACACCGGCAARTCCAT3') and LowerA (5'GC...
Embodiment 2
[0018] Example 2 Construction of Escherichia coli expression vector pLyase-A
[0019] Design upstream primers (5'CCGAAT TCA TCC CCC ACA GG GTT ACT AC G A3') and downstream primers (5'CCC AAGCTT GCT TTT TTG CTC TCT GCG TTC G3') according to the obtained complete sequence of the alginate lyase gene algL; The complete sequence of the alginate lyase gene was amplified by PCR. The PCR conditions were as follows: pre-denaturation at 94°C for 3 minutes, followed by 30 cycles of 94°C for 30s, 60°C for 30s, and 72°C for 60s, and finally extension at 72°C for 10 minutes. Agarose gel electrophoresis showed a specific band at 1.0 kb, which was excised from the agarose gel, digested with Hind III and EcoR I, and a 1.0 kb DNA fragment was recovered after agarose gel electrophoresis.
[0020] The Escherichia coli expression vector pBAD gIII / A was also digested with Hind III and EcoR I, separated by agarose electrophoresis, and a 4.1kb DNA fragment was recovered, which was connected to the a...
Embodiment 3
[0021] Example 3 Construction of Escherichia coli engineering strain pLyase-A / Top10F' capable of highly expressing alginate lyase
[0022] The expression vector pLyase-A was transformed into Escherichia coli Top10F' according to the standard calcium chloride method, and transformants with ampicillin resistance were screened. The plasmid was extracted by a standard alkaline lysis method, and two fragments of 1.0 kb and 4.1 kb were obtained by Hind III and EcoR I digestion, which were respectively combined with the full-length fragment of the alginate lyase gene algL and the expression vector pBAD gIII / A, proving that the energy Escherichia coli recombinant strain pLyase-A / Top10F' highly expressing alginate lyase.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com