Novel fungal proteins and nucleic acids encoding same
A protein, nucleic acid molecule technology, applied in the field of recombinant production of the nucleic acid and peptidase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0260] Example 1: Materials and methods
[0261] Strains and plasmids
[0262] A clinical isolate of Trichophyton rubrum, CHUV 862-00, was used in this study. Bacteriophage lambda EMBL3 (Promega, Wallisellen, Switzerland) was propagated with E. coli LE392. All plasmid-subcloning experiments were performed in E. coli DH5α using the plasmid pMTL2I (Chambers et al., Gene 68:139-149 (1988)). The recombinant peptidase was expressed using P. pastoris GSI15 and the expression vector pKJ 113 (Borg-Von Zepelin et al, Mol. Microbiol 28:543-554 (1998)). It is well known in the art that P. pastoris can be used to express a large number of recombinant proteins.
[0263] Trichophyton rubrum growth medium
[0264] Trichophyton rubrum was grown on Sabouraud agarose or liquid medium (Bio-Rad, Munchen, Germany), or in order to increase the proteolytic activity of the product, in a medium containing 0.2% soybean protein (Supro 1711, Protein Technology International, St. Lauise. MO) as the s...
Embodiment 2
[0308] Example 2: Hydrolase Activity of Trichophyton rubrum Secreted Proteins
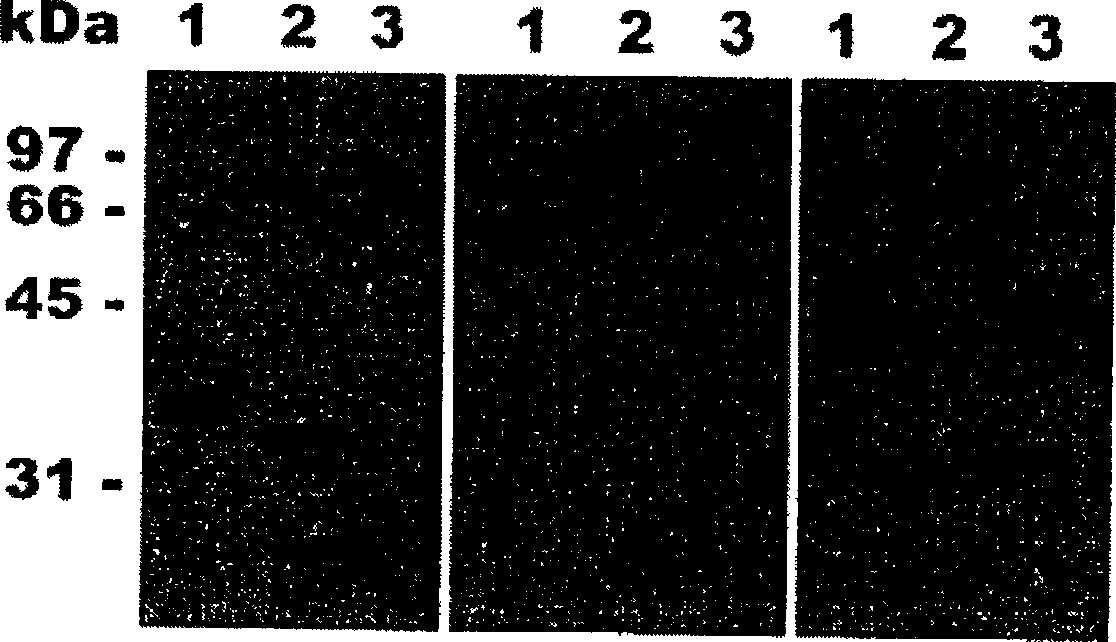
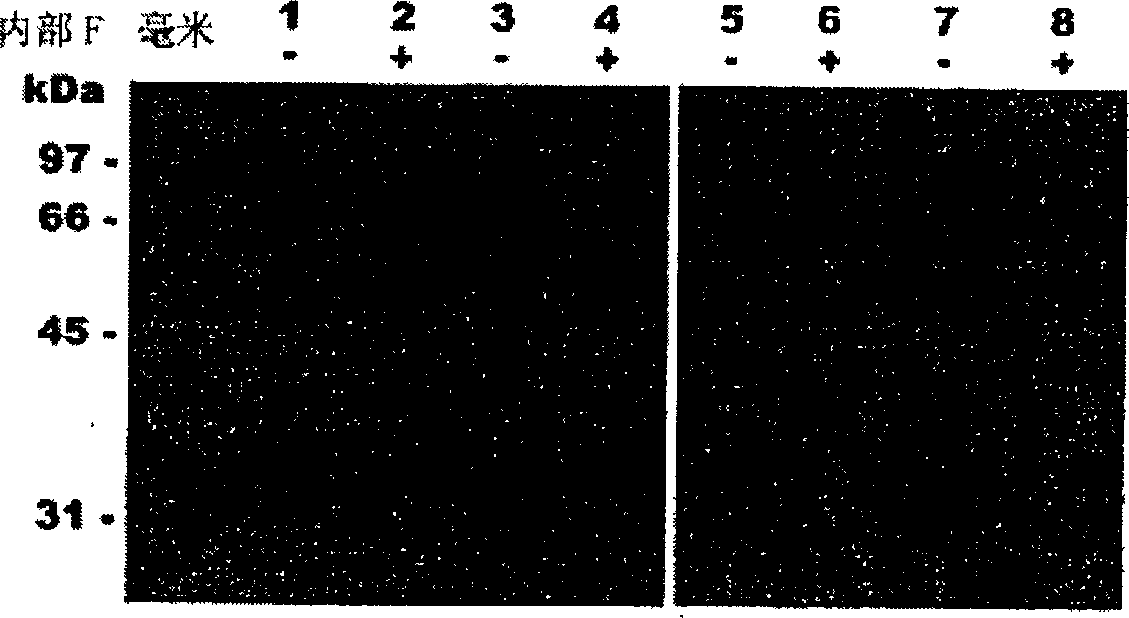
[0309] Trichophyton rubrum was cultured at 30°C on a medium containing 0.2% soybean protein as the sole nitrogen and carbon source. After 14 days of growth, accompanied by clarification of the medium was recorded, real proteolytic enzyme activity (400 U / ml) was detected using resorufin-labeled casein as substrate. Its proteolytic activity was inhibited by 15% and 85% by PMSF and o-phenanthroline, respectively, demonstrating that Trichophyton rubrum secretes serine and metalloproteases. Western blot analysis of the culture supernatant revealed that Trichophyton rubrum was similar to Microsporum canis, secreting endogenous proteases of the subtilisin family (MEROPS>S8) and fungalysin family (MEROPS>M36), similar to A .oryzae secretes alkaline phosphatase ALP and neutral metalloprotease NPI (see figure 1 ). In addition, high activities of substrates such as Leu-AMC and Leu-pNA were detected in the ...
Embodiment 3
[0310] Example 3: Aminopeptidase activity of Trichophyton rubrum secretions
[0311] The nucleotide sequence of the M. canis endoprotease gene is 50-70% similar to the homologous gene. These genes encode subtilisin and fungalysin secreted by A. oryzae and A. fumigtus. In addition, the genes of M. canis and Aspergillis have a linear correspondence of intron-exon structure. Therefore, the DNA sequences of genes encoding aminopeptidases from A. oryzae and yeast were used to design probes for screening T. rubrum genomic DNA libraries. Using the recombinant protein, by comparison with the protein secreted by the opportunistic pathogen Aspergillus fumigatus. Characterization of aminopeptidase secreted by Trichophyton rubrum.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com