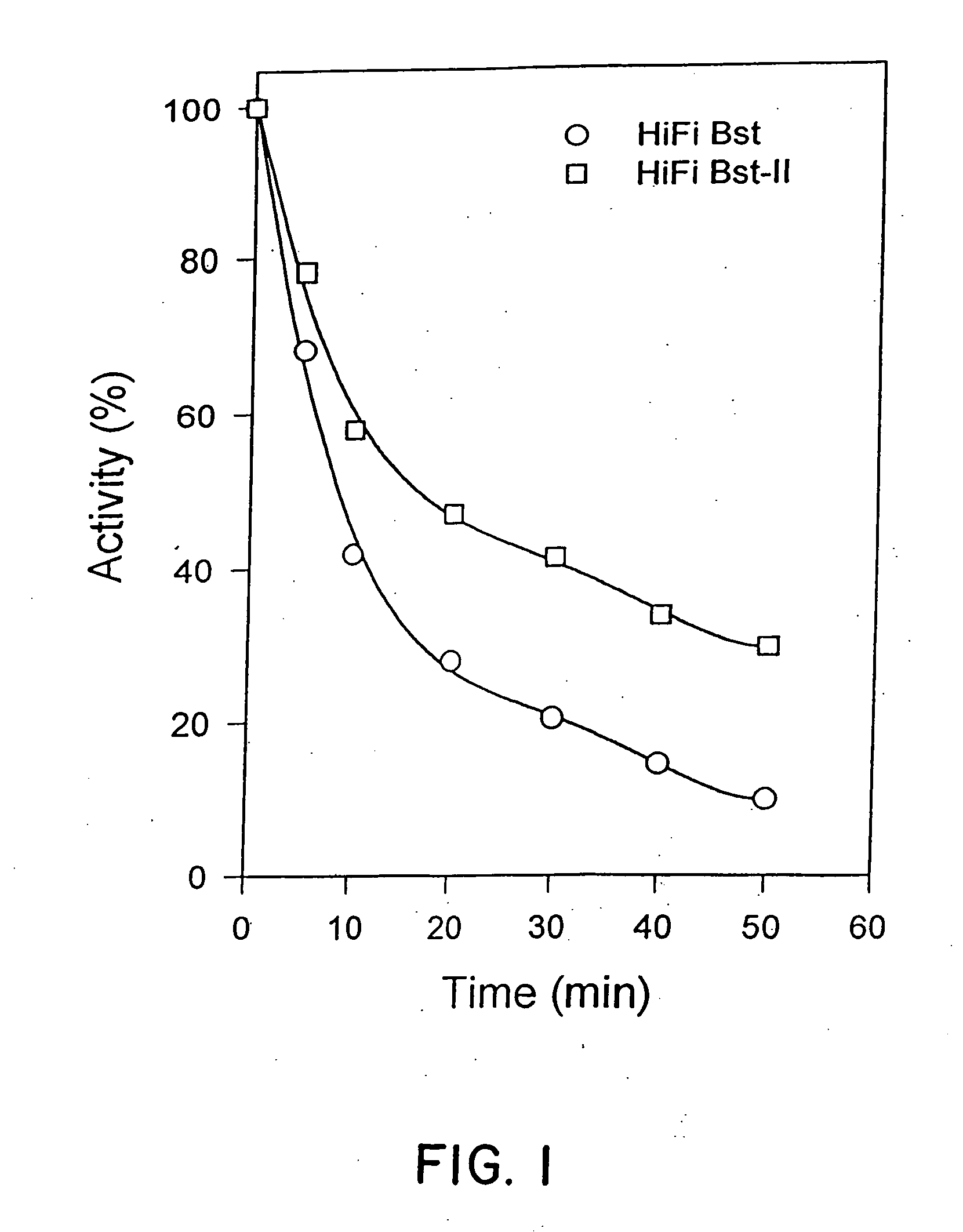
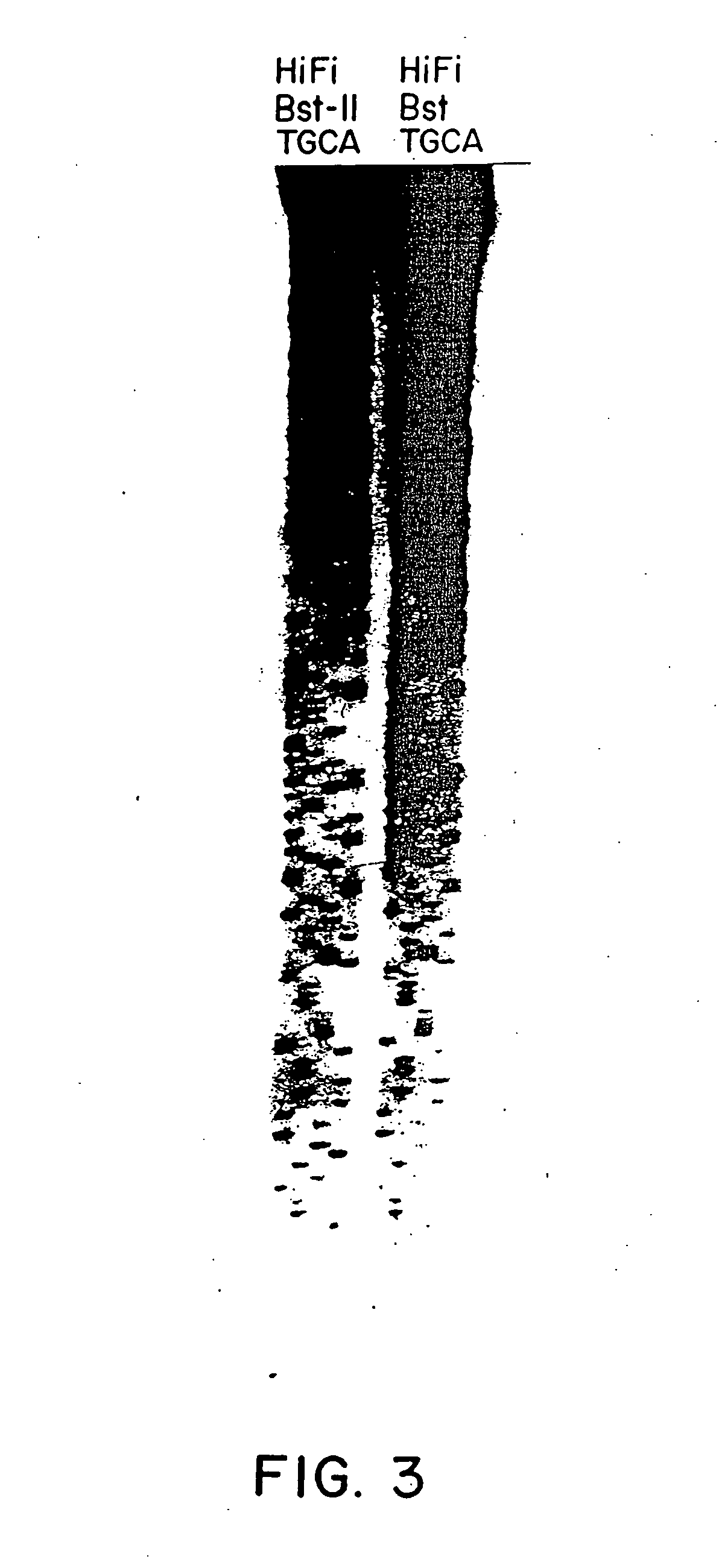DNA polymerase having ability to reduce innate selective discrimination against fluorescent dye-labeled dideoxynucleotides
a technology of dideoxynucleotide and dna polymerase, which is applied in the field of dna polymerase having ability to reduce the innate selective discrimination of fluorescent dye-labeled dideoxynucleotides, can solve the problems of many undesirable, low processivity, and generation of erroneous or misleading band patterns of radioactivity in the sequencing gel, so as to reduce the innate selective discrimination, and increase the rate of incorpor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Screening for Bst Polymerases
[0106] This invention also involves a method to measure the proof-reading 3′-5′ exonuclease activity of purified DNA polymerases. The method is useful to screen a large number of bacterial strains, such as Bacillus stearothermophilus and other mesophilic bacterial strains, to select a strain which produces a DNA polymerase with a high proof-reading 3′-5′ exonuclease activity. For instance, the method to test the proof-reading 3′-5′ exonuclease activity of DNA polymerase was carried out as follows.
[0107] A DNA primer and two DNA templates with following sequences were synthesized chemically, using a DNA synthesizer.
17-base primer 5′ CATTTTGCTGCCGGTCA 3′(SEQ ID NO: 5) 1 mg / mlTemplate (a)3′------GTAAAACGACGGCCAGTCTT------5′(SEQ ID NO: 6) 10 mg / mlTemplate (b) 3′-----GTAAAACGACGGCCAGTCGG-----5′(SEQ ID NO: 7) 10 mg / ml
[0108] To produce the radiolabeled primer, 1 μl (1 μg) of primer, 5 μl (50 μg) of templat...
example 2
Mutation of the Gene for Naturally-Occurring Bst DNA Polymerase Having Proofreading 3′-5′ Exonuclease Activity
[0121] The DNA fragment LF containing the gene initially isolated from the wild Bst 320 has the following sequence (see SEQ ID NO:1):
[0122] DNA Sequence (Isolated / Purified):
GCCGAAGGGG AGAAACCGCT TGAGGAGATG GAGTTTGCCATCGTTGACGT CATTACCGAA GAGATGCTTG CCGACAAGGCAGCGCTTGTC GTTGAGGTGA TGGAAGAAAA CTACCACGATGCCCCGATTG TCGGAATCGC ACTAGTGAAC GAGCATGGGCGATTTTTTAT GCGCCCGGAG ACCGCGCTGG CTGATTCGCAATTTTTAGCA TGGCTTGCCG ATGAAACGAA GAAAAAAAGCATGTTTGACG CCAAGCGGGC AGTCGTTGCC TTAAAGTGGAAAGGAATTGA GCTTCGCGGC GTCGCCTTTG ATTTATTGCTCGCTGCCTAT TTGCTCAATC CGGCTCAAGA TGCCGGCGATATCGCTGCGG TGGCGAAAAT GAAACAATAT GAAGCGGTGCGGTCGGATGA AGCGGTCTAT GGCAAAGGCG TCAAGCGGTCGCTGCCGGAC GAACAGACGC TTGCTGAGCA TCTCGTTCGCAAAGCGGCAG CCATTTGGGC GCTTGAGCAG CCGTTTATGGACGATTTGCG GAACAACGAA CAAGATCAAT TATTAACGAAGCTTGAGCAC GCGCTGGCGG CGATTTTGGC TGAAATGGAATTCACTGGGG TGAACGTGGA TACAAAGCGG CTTGAACAGATGGGTTCGGA GCTCGCCGAA ...
example 3
Cloning and Expression of the Modified Bst DNA Polymerase Having Both Ability to Reduce Selective ddNTP Discrimination and Proofreading 3′-5′ Exonuclease Activity
[0134] The plasmid pUC119 / LF-M was prepared from the strain of Escherichia coli JM109 containing the mutated DNA. The mutated DNA fragment (LF-M) containing the mutated gene for the Bst polymerase was recombined back into the expression vector pYZ23. The constructed plasmid pYZ23 / LF-M was then transformed into Escherichia coli JF1125. The mutation was further confirmed by double-stranded dideoxy DNA sequencing of isolated plasmid.
[0135] The strain of Escherichia coli JF1125 containing pYZ23 / LF-M was inoculated into LB culture containing 100 μg / ml ampicillin, and was incubated overnight at 30° C. The overnight culture was inoculated into a large volume of fresh culture, and was incubated at 30° C. until the OD600 of the culture reached 0.7. The culture was then heated at 41° C. for 3 hours for induction. The SDS-PAGE analy...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com