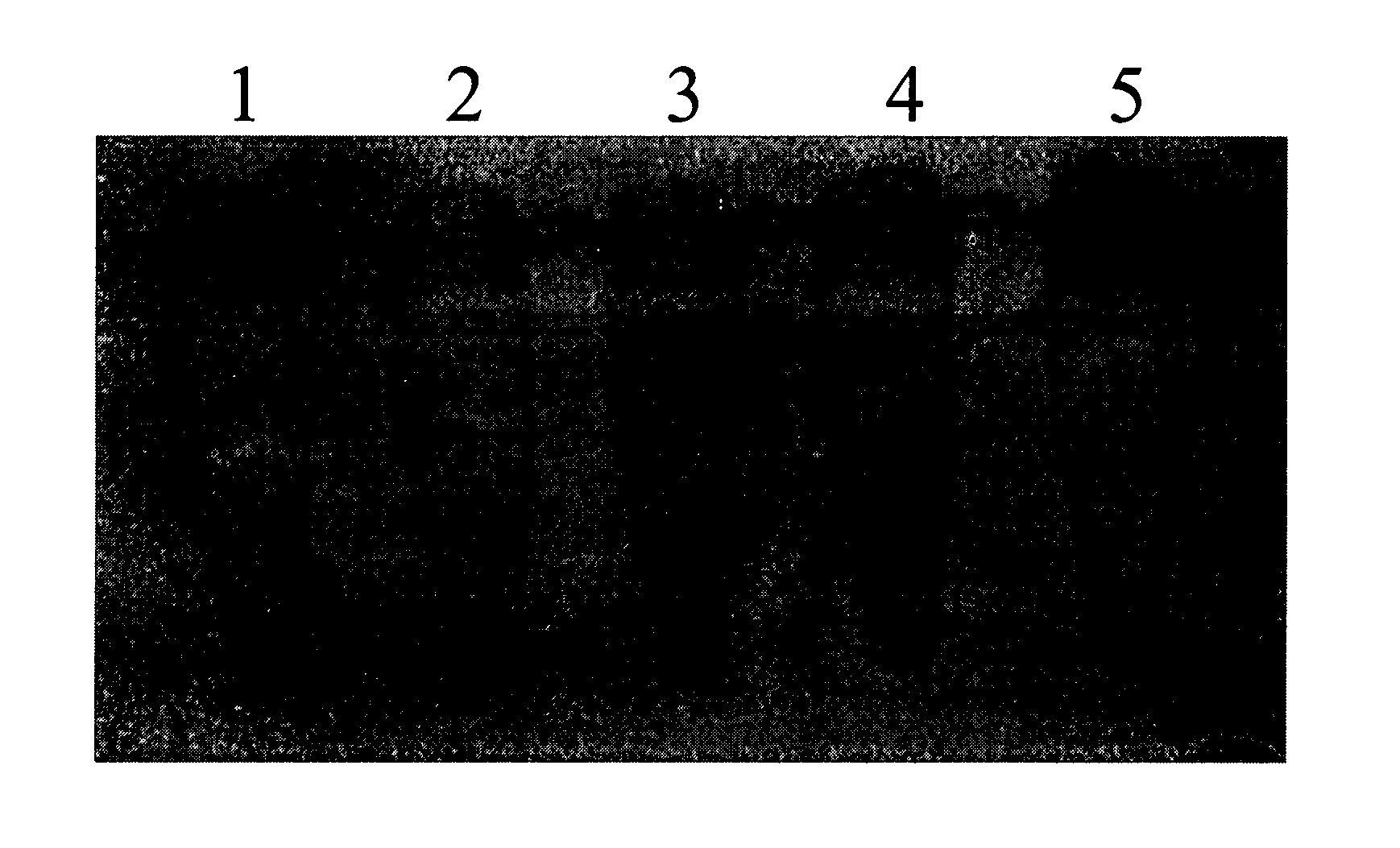
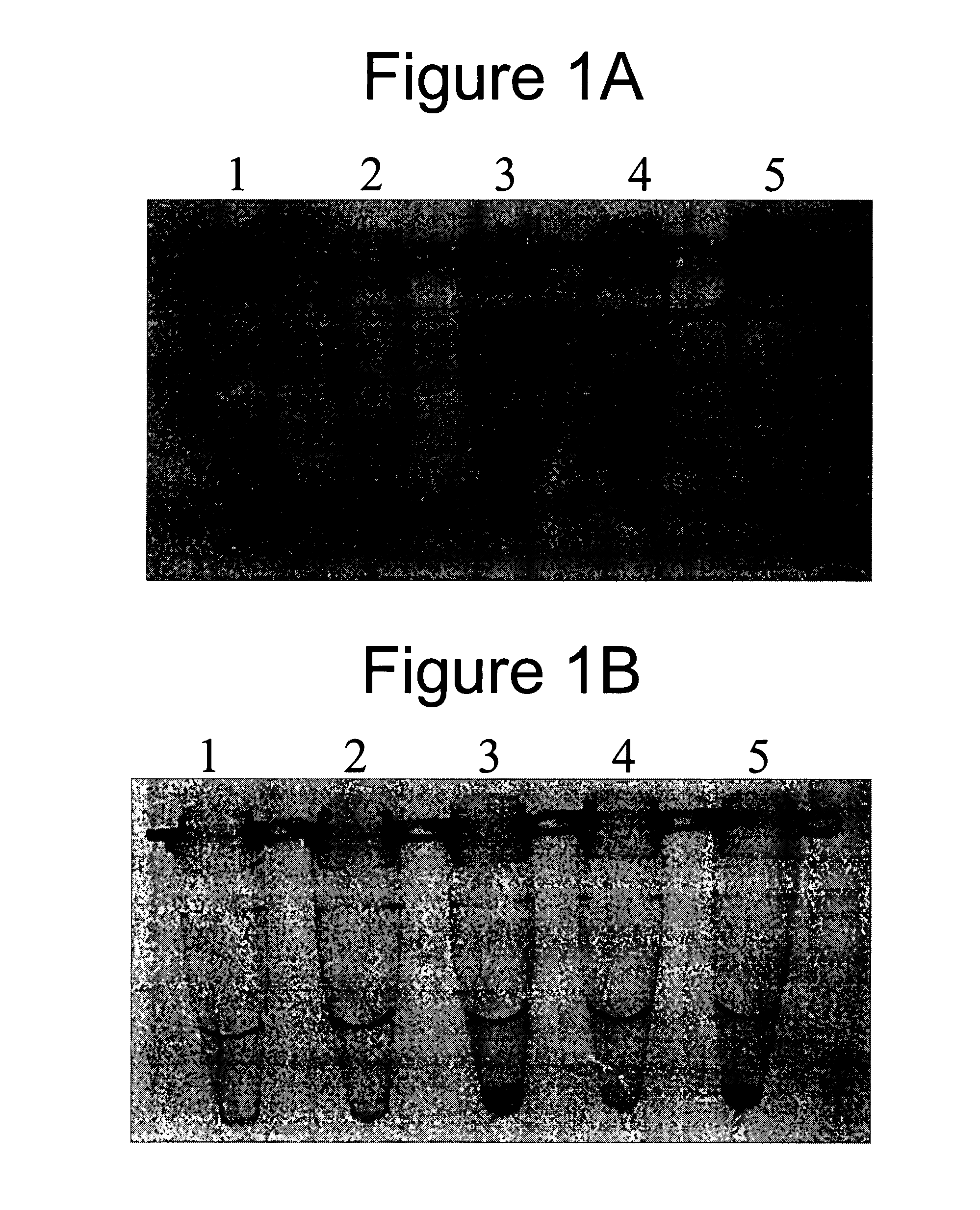
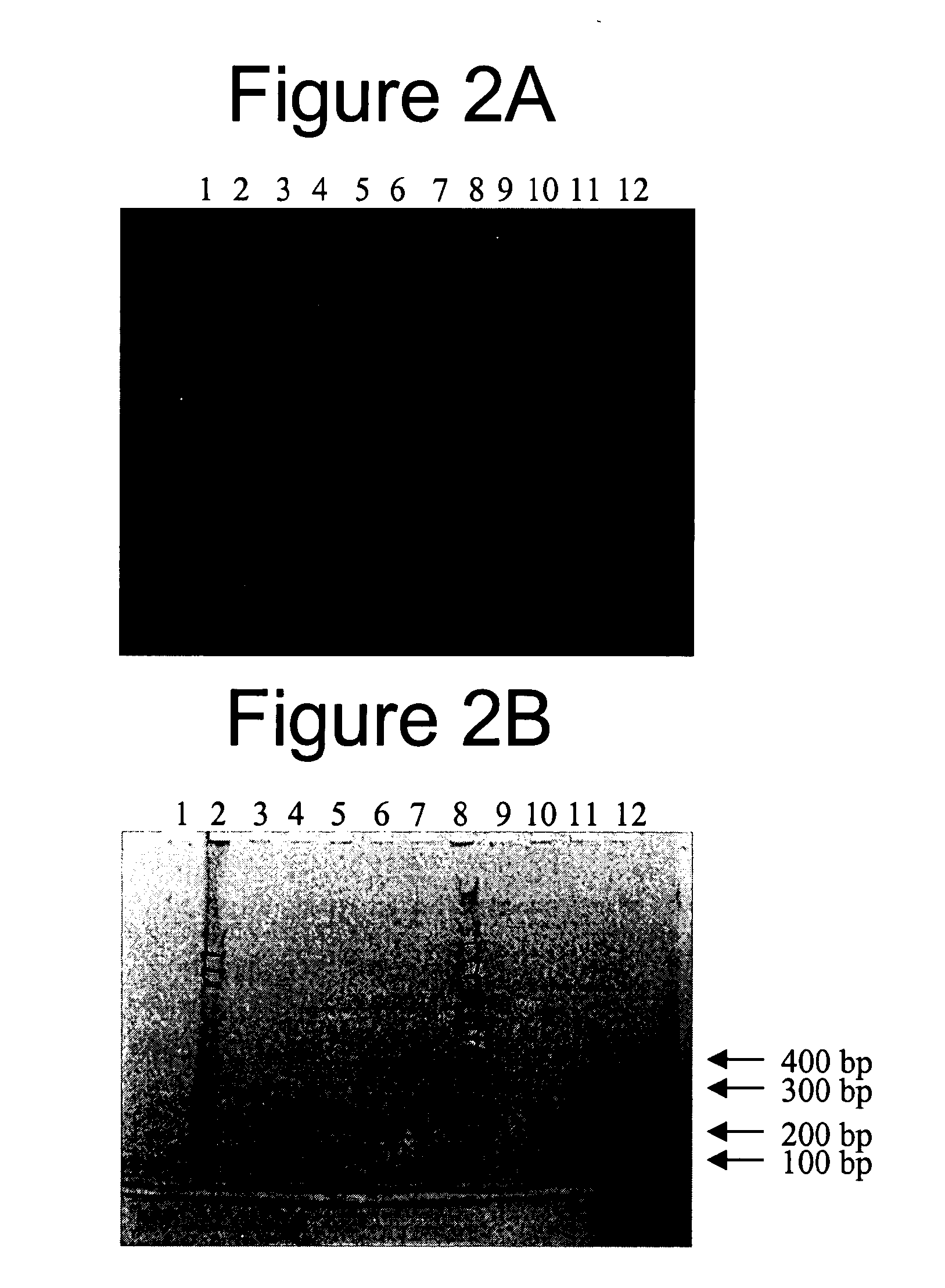
Methods, Kits and Compositions for the Identification of Nucleic Acids Electrostatically Bound to Matrices
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of DNA Oligonucleotides for Study
[0117]For this study, labeled and labeled DNA oligonucleotides suitable as probes or as nucleic acids comprising a target sequence were either synthesized using commercially available reagents and instrumentation or obtained from commercial vendors. All DNAs were obtained in purified form or purified using conventional methods. The sequences of the DNA oligonucleotides prepared are illustrated in Table 1, below. Methods and compositions for the synthesis and purification of synthetic DNAs are well known to those of ordinary skill in the art.
example 2
Synthesis of N-α-(Fmoc)-N-ε-(NH2)-L-Lysine-OH
[0118]To 20 mmol of N-α-(Fmoc)-N-ε-(t-boc)-L-lysine-OH was added 60 mL of 2 / 1 dichloromethane (DCM) / trifluoroacetic acid (TFA). The solution was allowed to stir until the tert-butyloxycarbonyl (t-boc) group had completely been removed from the N-α-(Fmoc)-N-ε-(t-boc)-L-lysine-OH. The solution was then evaporated to dryness and the residue redissolved in 15 mL of DCM. An attempt was then made to precipitate the product by dropwise addition of the solution to 350 mL of ethyl ether. Because the product oiled out, the ethyl ether was decanted and the oil put under high vacuum to yield a white foam. The white foam was dissolved in 250 mL of water and the solution was neutralized to pH 4 by addition of saturated sodium phosphate (dibasic). A white solid formed and was collected by vacuum filtration. The product was dried in a vacuum oven at 35-40° C. overnight. Yield 17.6 mmol, 88%.
example 3
Synthesis of N-α-(Fmoc)-N-ε-(dabcyl)-L-Lysine-OH
[0119]To 1 mmol of N-α-(Fmoc)-N-ε-(NH2)-L-Lysine-OH (Example 2) was added 5 mL of N,N′-dimethylformamide (DMF) and 1.1 mmol of TFA. This solution was allowed to stir until the amino acid had completely dissolved.
[0120]To 1.1 mmol of 4-((4-(dimethylamino)phenyl)azo)benzoic acid, succinimidyl ester (Dabcyl-NHS; Molecular Probes, P / N D-2245) was added 4 mL of DMF and 5 mmol of diisopropylethylamine (DIEA). To this stirring solution was added, dropwise, the N-α-(Fmoc)-N-ε-(NH2)-L-Lysine-OH solution prepared as described above. The reaction was allowed to stir overnight and was then worked up.
[0121]The solvent was vacuum evaporated and the residue partitioned in 50 mL of DCM and 50 mL of 10% aqueous citric acid. The layers were separated and the organic layer washed with aqueous sodium bicarbonate and again with 10% aqueous citric acid. The organic layer was then dried with sodium sulfate, filtered and evaporated to an orange foam. The foam...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap