Methods for predicting risk of metastasis in cutaneous melanoma
a technology for cutaneous melanoma and risk prediction, which is applied in the direction of microbiological testing/measurement, biochemistry apparatus and processes, peptide/protein ingredients, etc., can solve the problems of inaccurate diagnosis, patient and doctor false sense of security about the extent of cancer in the patient's body, and inaccurate prognosis, etc., to achieve enhanced surveillance, accurate and objective, and low positivity rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cutaneous Melanoma Metastatic Risk Genetic Signature and Biomarker Expression
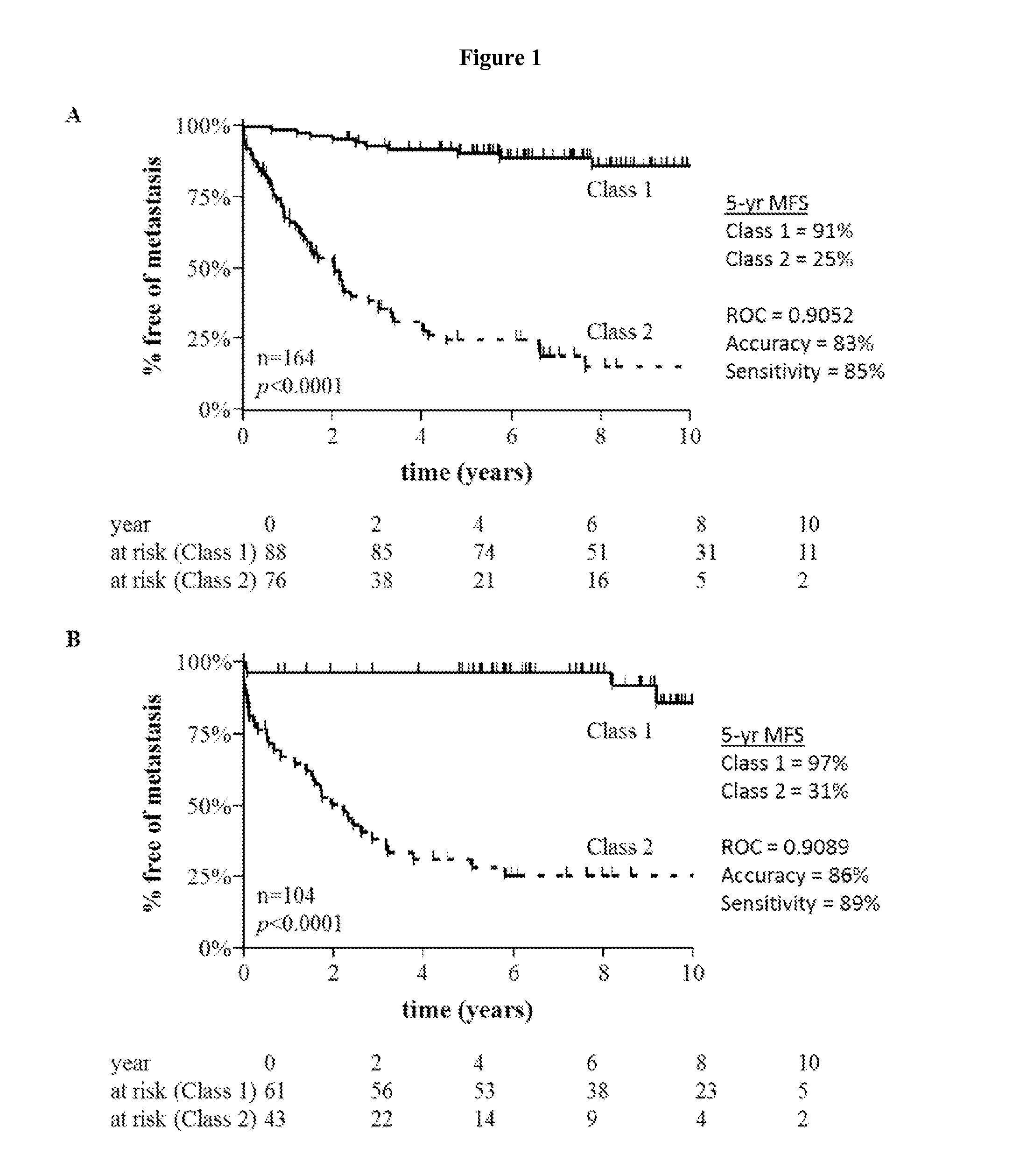
[0064]Genetic expression of the discriminant genes in the signature (Table 1) was assessed in a cohort of 268 cutaneous melanoma samples using RT-PCR (FIG. 1). As shown in Table 2 below, of the 28 discriminating genes, 26 were significantly altered in metastatic melanoma tumors compared to nonmetastatic tumors (p<0.05, range 0.0366-6.08E-16), and 25 were down-regulated. Genes that were up-regulated in the metastatic tumors included SPP1, KRT6B, and EIF1B.
TABLE 1Genes included in the GEP signature able to predict metastatic risk from primary CM tumors.Gene SymbolGene NameAlternative Gene NamesBAP1_varABRCA1 associated protein-1TPDS, UCHL2, HUCEP-13, HUCEP-6,BAP_var1, BAP (a1)BAP1_varBBRCA1 associated protein-1UCHL2, HUCEP-13, HUCEP-6, BAP_var2,BAP (a2)MGPmatrix gamma-carboxyglutamicmatrix Gla protein, gamma-carboxyglutamicacidacid, GIG36, MGLAP, NTISPP1secreted phosphoprotein 1BNSP, BSPI, ETA-1, OPN, PSEC015...
example 2
Initial Training Set Development Studies and Comparison to Validation Cohort
[0065]Using JMP GENOMICS® software and clinical data analysis, a training set of 164 cutaneous melanoma samples was generated that could accurately predict the risk of metastasis based upon the 28 gene signature. The training set contained 15 stage 0 in situ melanomas, 61 stage I, 70 stage II, 17 stage III, and 1 stage IV melanomas. Metastatic risk was assessed using a radial basis machine predictive modeling algorithm, which reports class 1 (low risk of metastasis) or class 2 (high risk of metastasis). ΔCt values generated from RT-PCR analysis of the training set cohort were standardized to the mean for each gene, with a scale equivalent to the standard deviation. Analyses were also performed using KNN, PTA, and discriminant analysis to confirm the results from the RBM approach (as discussed below). The training set prediction algorithm was then validated using an independent cohort of 104 cutaneous melanom...
example 3
Analysis of 162 Sample Training Set with Multiple Predictive Modeling Methods
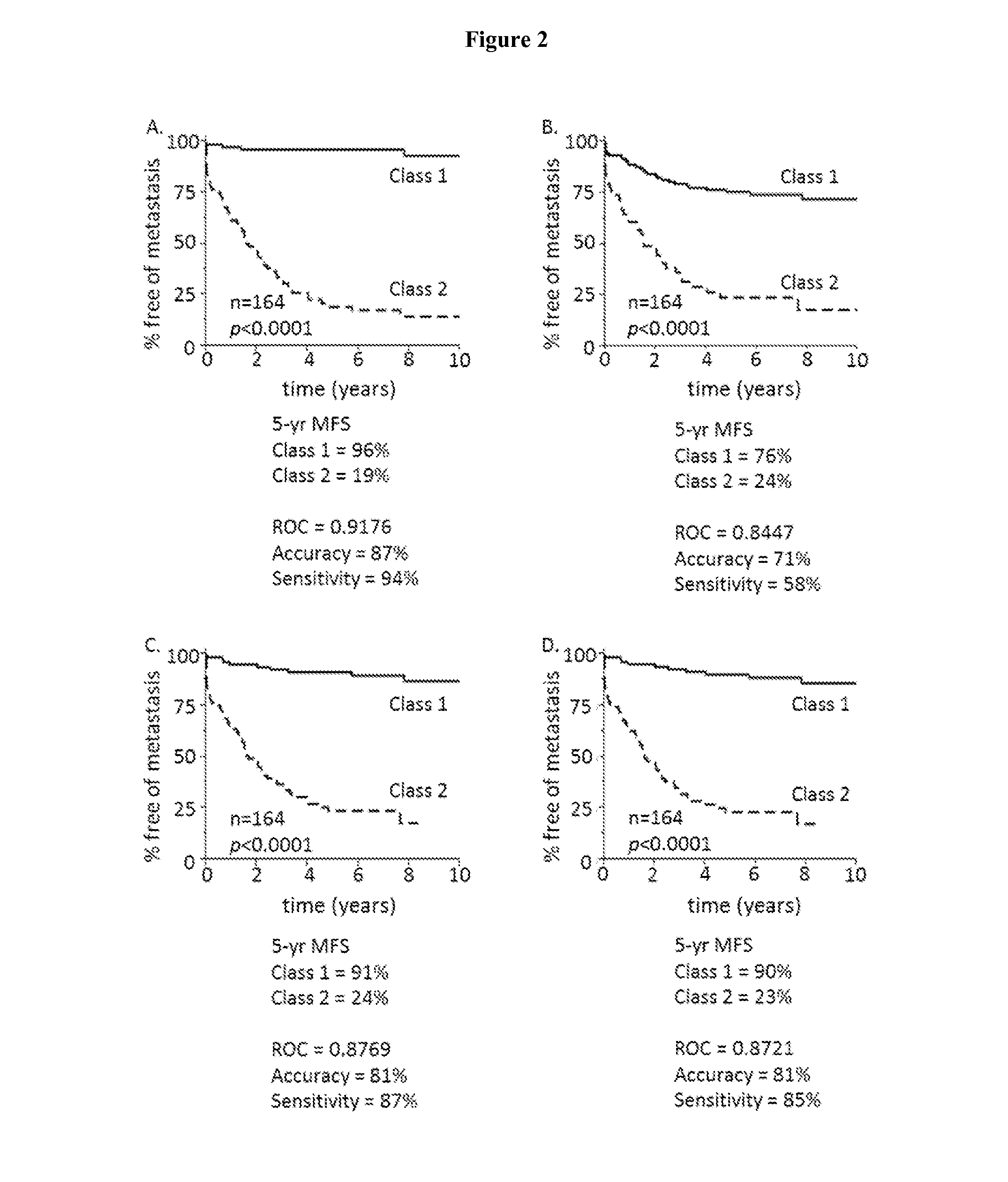
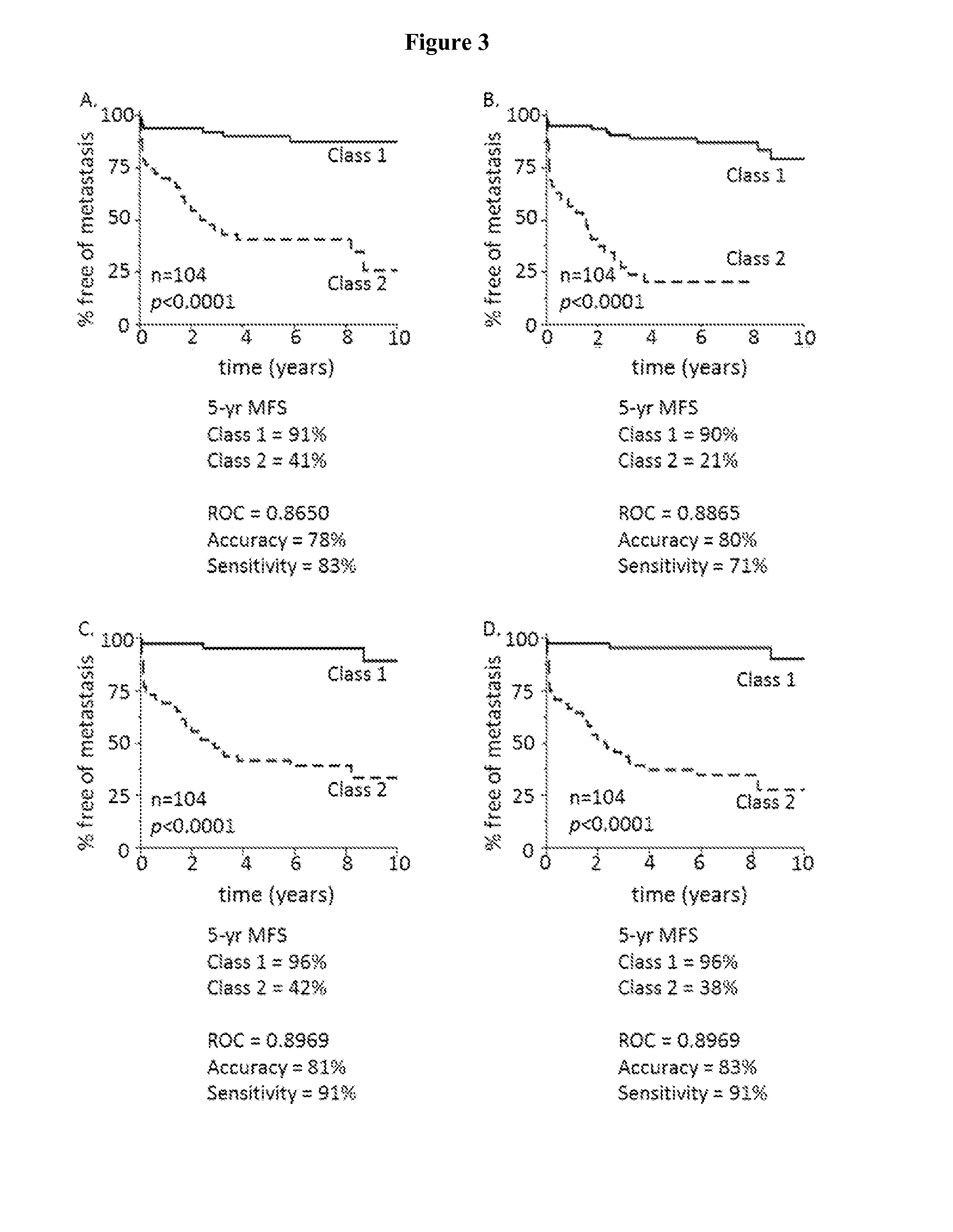
[0067]JMP GENOMICS® software allows for analysis using linear and non-linear predictive modeling methods. To assess whether accuracy of metastatic risk prediction for the validation cohort was limited to the RBM method, partition tree, K-nearest neighbor, logistic regression, and discriminant analysis were performed (FIGS. 2 and 3). Training set ROC, accuracy, sensitivity, specificity and K-M 5-year MFS for class 1 and class 2 cases were highly comparable to the RBM method. Highly accurate prediction of metastasis, and significantly different 5-year MFS between class 1 and class 2 cases was observed when using partition tree (FIG. 2A), K-nearest neighbor (FIG. 2B), logistic regression (FIG. 2C), or discriminant analysis (FIG. 2D). Significant differences in 5-year MFS were also observed for validation cohort class 1 and class 2 cases using partition tree (FIG. 3A), K-nearest neighbor (FIG. 3B), logistic reg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com